Fig. 3. Polymethylation score analyses on disease risk and patient survival.
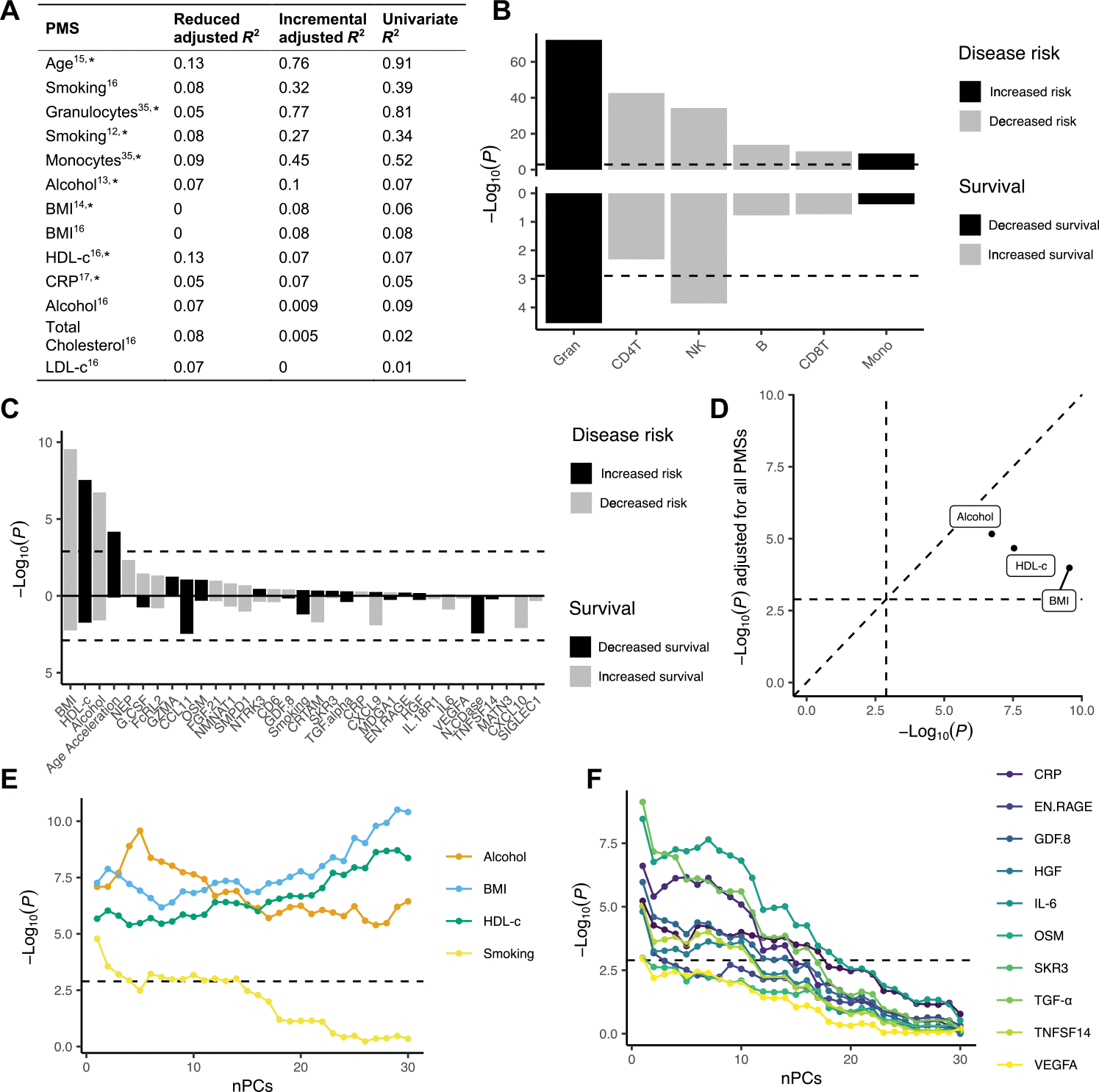
Polymethylation scores (PMSs) were determined as proxies for various traits, exposures, proteins, and WBC proportions, calculated as weighted sums based on probes and weights derived from published papers, respectively. Case-control association analyses were performed on 6763 patients and 2943 controls; survival analyses were performed within 5162 patients. (A) Explained variance of PMSs calculated in samples for which both DNA methylation data and biomarker/clinical data were available (N = 800 of 2000). Reduced R2 represents the variance explained by the null model, whereas the incremental R2 represents the additional variance explained by the PMS over the null model. Last, the explained variance of the univariate model of the respective PMS is displayed (see Materials and Methods). The asterisk indicates that the PMS was used in the association tests. (B and C) The top panel shows association P values from logistic regression [−log10(P), y axis] for each PMS (x axis). (B) WBC proportions and (C) various traits and exposures, colored by whether a higher score is associated with increased (black) or decreased (gray) disease risk. The bottom panel shows the Cox PH P values [−log10(P), y axis] for each PMS (x axis), colored by whether a higher score is associated with decreased (black) or increased (gray) survival, respectively. The dashed line indicates the significance threshold (1.3 × 10−3). (D) Original P values [−log10(P), x axis] compared to P values after including all PMSs as fixed covariates in the logistic regression model [−log10(P), y axis] for the ALS-associated traits/exposures. (E and F) Association P values [−log10(P), y axis] upon incrementally adding principal components (PCs) as fixed covariates in the logistic regression model. HGF, hepatocyte growth factor; EN.RAGE, extracellular newly identified RAGE-binding protein; GDF8, growth/differentiation factor 8; OSM, Oncostatin-M, SKR3, Serine/threonine-protein kinase receptor R3; TNFSF14, tumor necrosis factor ligand superfamily member 14; VEGFA, vascular endothelial growth factor A; nPCs, number of principal components.
