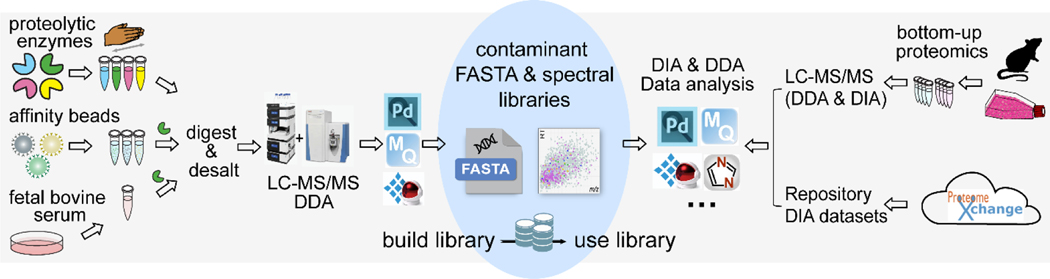
Figure 1: Schematic of building and using the contaminant libraries for DDA and DIA proteomics.
A series of contaminant-only samples were created by adding different proteolytic enzymes to keratin-contaminated lysis buffer, commonly used beads coated with affinity tags, and fetal bovine serum (FBS) for cell culture medium. New contaminant FASTA and spectral libraries were created using DDA proteomic analyses of contaminant-only samples. These new contaminant libraries were evaluated using different biological samples and repository datasets in various DDA and DIA software platforms.