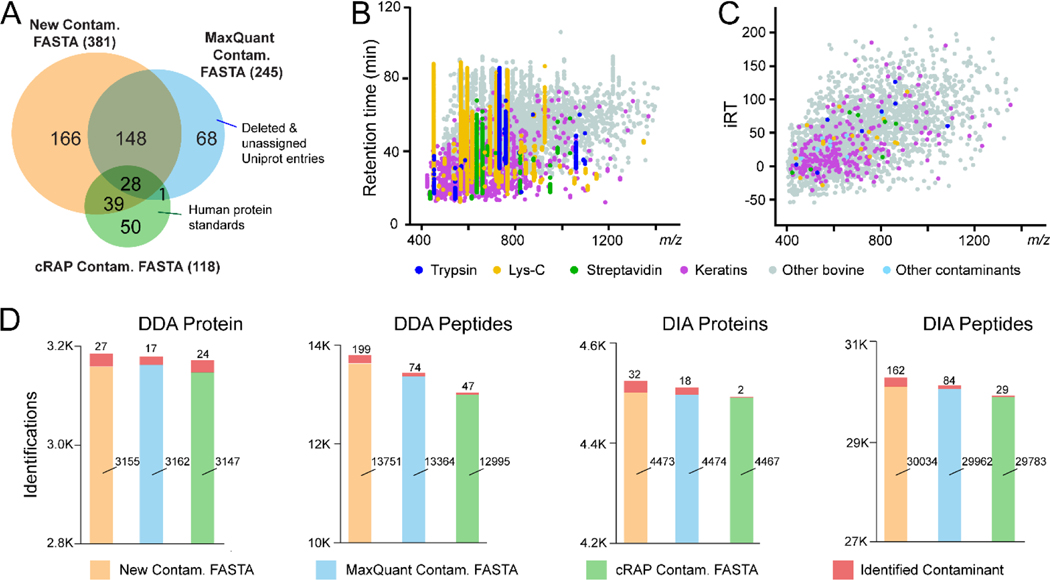
Figure 2: Characterization of the contaminant protein FASTA and spectral libraries.
(A) Venn diagram comparison of contaminant protein lists from our newly generated contaminant FASTA and commonly used MaxQuant and cRAP contaminant FASTA files. (B) Scatterplot of identified contaminant peptides merged from contaminant-only samples in DDA LC-MS/MS analyses. (C) Scatterplot of contaminant peptides in our contaminant spectral libraries, generated by the Spectronaut Pulsar. iRT stands for in-silico normalized retention time. (D) Comparison of DDA and DIA protein and peptide identifications from HEK samples using our new contaminant FASTA in comparison to the MaxQuant and cRAP FASTA libraries.