Fig. 1.
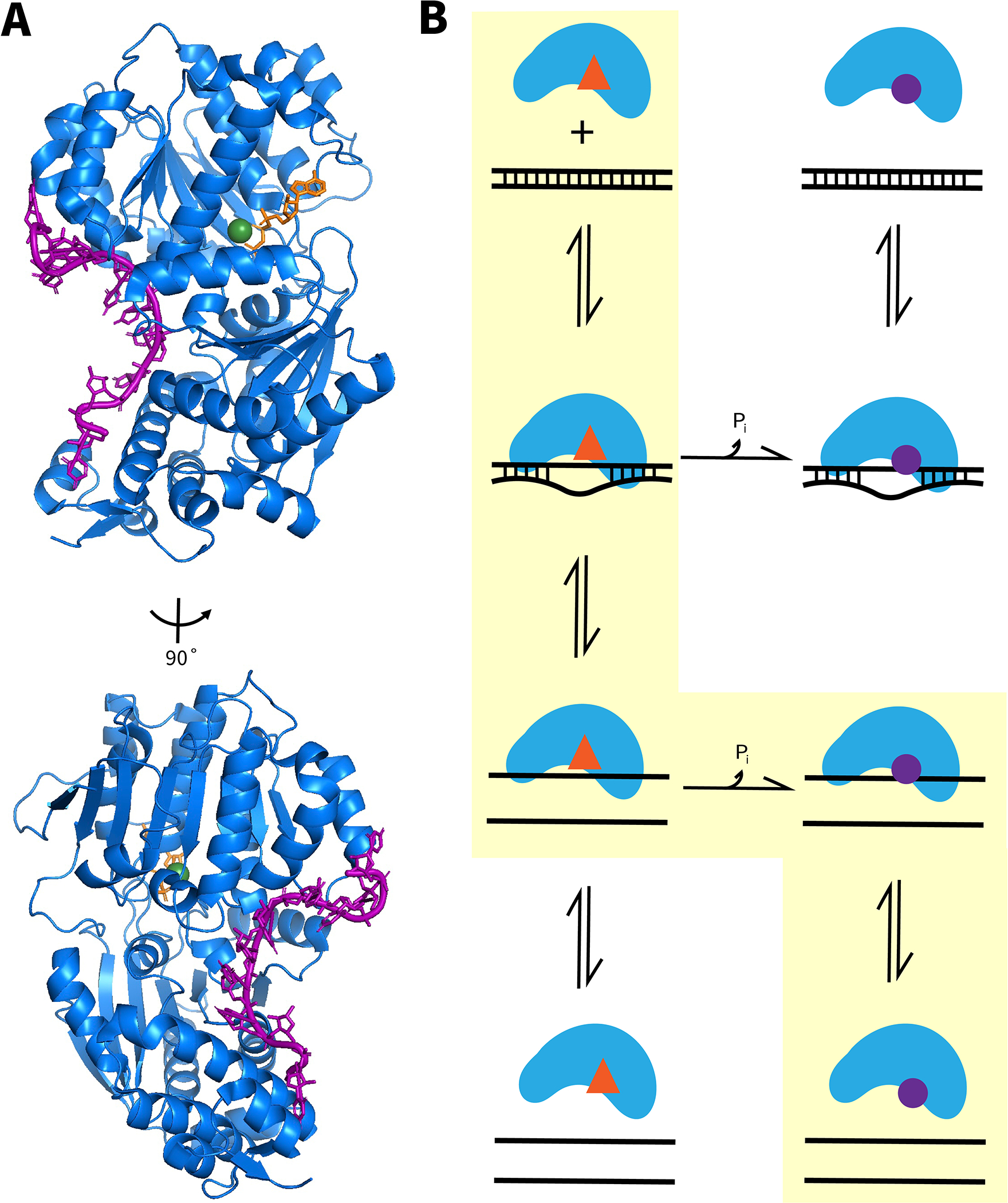
RNA binding and unwinding by DEAD-box helicase proteins. (A), Structural views of ssRNA binding by the helicase core, as shown for Mss116 (Del Campo & Lambowitz, 2009b). The ssRNA (purple) binds to Mss116 (blue) in a crimped conformation, incompatible with a standard duplex geometry. Also shown are a bound ATP analog (orange) and a coordinated Mg2+ ion (green). (B) ATP-dependent RNA unwinding by DEAD-box helicases. In the dominant pathway, highlighted in yellow, the ATP-bound form of the helicase binds dsRNA and produces local unwinding (starting from top left; ATP is indicated by a red triangle). This unwinding reaches completion, resulting in the dissociation of one of the strands, producing the ssRNA-bound form shown in panel A. ATP hydrolysis and release of Pi weaken binding of ssRNA (bound ADP is indicated by a purple circle), resulting in ssRNA dissociation (bottom right). Thus, this pathway results in complete helix unwinding with hydrolysis of one ATP. Less populated pathways produce ATPase activity without complete duplex unwinding (top two rows, counterclockwise) and complete duplex unwinding without ATP hydrolysis (left column, top to bottom).
