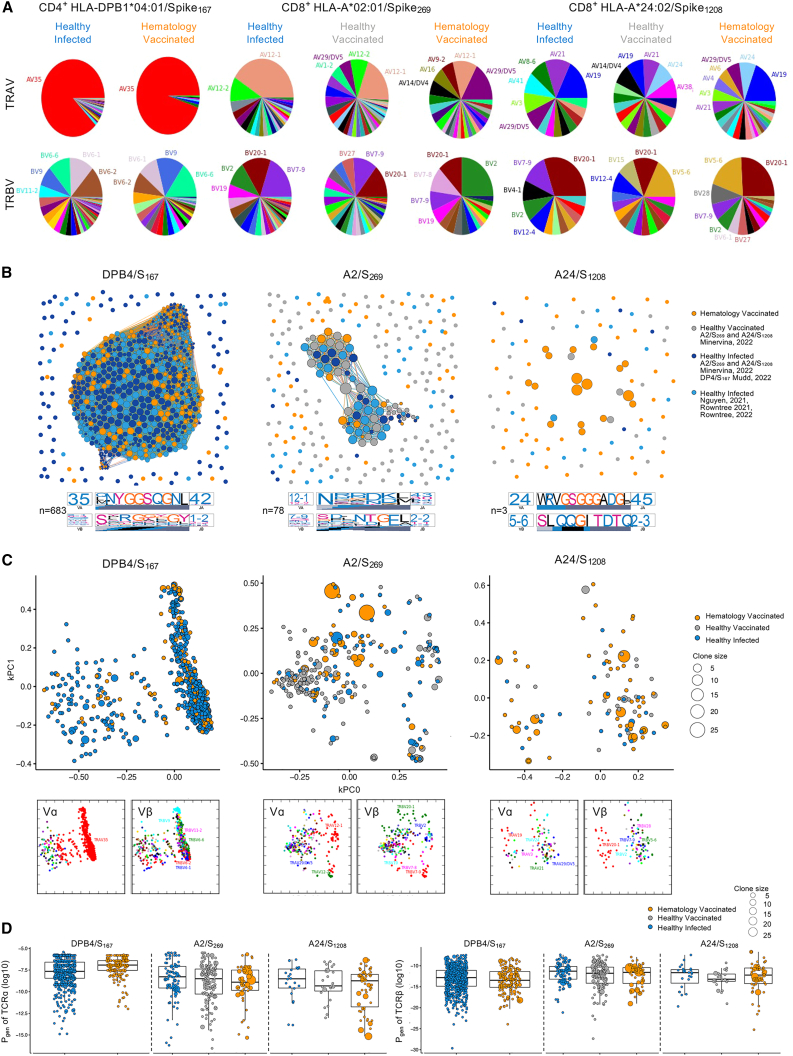
Figure 6.
TCR sequence similarity network identifies sharing of dominant motifs between the groups
(A) Pie charts of TRAV and TRBV usage for TCRαβ clonotypes specific to DPB4/S167 (n = 18 patients), A2/S269 (n = 18 patients), and A24/S1208 (n = 13 patients). Clonally expanded TCRs were reduced to a single data point for this analysis.
(B) TCRαβ sequence similarity networks. Each vertex is a different clonotype, and edges connect clonotypes with highly similar amino acid TCRαβ sequences (TCR distance [TCRdist] ≤ 120). Size of the vertex is proportional to number of neighbors. TCRdist sequence logos for TCRα and TCRβ chains are shown for central largest cluster for each panel. Each TCR motif depicts the V (left side) and J (right side) gene frequencies, the CDR3 amino acid sequence (middle), and the inferred rearrangement structure (bottom bars colored by source region; V-region, light gray; insertions, blue; diversity [D]-region, black; and J-region dark gray).
(C) TCR landscapes displayed using kernel principal-component analysis (PCA) projections. Vα/Vβ usage shown below.
(D) Pgen for TCRα and TCRβ. Boxplots represent the median (middle bar), 75% quantile (upper hinge), and 25% quantile (lower hinge), with whiskers extending 1.5 times the IQR.
Experiments were performed once for each sample. Refer to Figures S10 and S11.