Abstract
Muscle fibers are multinucleated, and muscle fiber nuclei (myonuclei) are believed to be post-mitotic and are typically situated near the periphery of the myofiber. Due to the unique organization of muscle fibers and their nuclei, the cellular and molecular mechanisms regulating myofiber homeostasis in unstressed and stressed conditions (e.g., exercise) are unique. A key role myonuclei play in regulating muscle during exercise is gene transcription. Only recently have investigators had the capability to identify molecular changes at high resolution exclusively in myonuclei in response to perturbations in vivo. The purpose of this review is to describe how myonuclei modulate their transcriptome, epigenetic status, mobility and shape, and microRNA expression in response to exercise in vivo. Given the relative paucity of high-fidelity information on myonucleus-specific contributions to exercise adaptation, we identify specific gaps in knowledge and provide perspectives on future directions of research.
Keywords: Skeletal muscle, Epigenetics, Muscle memory, Transcription, myomiR
Abbreviations
- Satellite cells
(SCs)
- Fibro/adipogenic progenitors
(FAPs)
- Ribonucleic acid
(RNA)
- Ribosomal RNA
(rRNA)
- Synergist ablation-induced mechanical overload
(SA)
- Progressive weighted wheel running
(PoWeR)
- Resistance exercise
(RE)
- Endurance exercise
(EE)
- Deoxyribonucleic acid
(DNA)
- MYC proto-oncogene
(Myc)
- RNA-sequencing
(RNA-seq)
- Extracellular matrix
(ECM)
- Rho guanosine triphosphate hydrolyze enzyme
(Rho-GTPase)
- Myosin heavy chain
(MyHC)
- Tumor necrosis factor
(TNF)
- TNF-like weak inducer of apoptosis
(TWEAK)
- TNF receptor superfamily member 12A
(Tnfrsf12a)
- Fibroblast growth factor-inducible 14
(Fn14)
- Wingless-related integration site
(Wnt)
- Nuclear Factor Kappa B
(NF-κB)
- Assay for transposase-accessible chromatin using sequencing
(ATAC-seq)
- Myostatin
(Mstn)
- Insulin-like growth factor 1
(IGF1)
- Mechano-growth factor
(MGF)
- Matrix metallopeptidase 9
(MMP9)
- Peroxisome proliferator-activated gamma coactivator-1 alpha
(PGC-1α)
- Peroxisome proliferator-activated receptor gamma
(PPAR-δ)
- Pyruvate dehydrogenase kinase 4
(PDK4)
- Kilodaltons
(kDa)
- Electrical pulse stimulation
(EPS)
- MicroRNAs
(miRNAs or miRs)
- Myofiber enriched microRNAs
(myomiRs)
- Phosphoinositide 3-kinase
(PI3k)
- Protein Kinase B
(AKT)
- Glucose-6-phosphate dehydrogenase
(G6pdx)
- Histone 2B- Green Fluorescent Protein
(H2B-GFP)
- Single myonucleus RNA sequencing
(smnRNA-seq)
Background
Skeletal muscle comprises approximately 40% of body mass in adult humans and plays an integral role in whole body energy metabolism, glucose homeostasis, and locomotion.1 Due to its abundance and central role in human health, biomedical research elucidating underlying mechanisms regulating muscle mass and function is of great importance. It is well established that exercise regulates the maintenance of healthy skeletal muscle throughout the lifespan.2,3 Regular physical activity reduces the risk of chronic disease, thus lowering burden on the health care system.4, 5, 6, 7, 8 Skeletal muscle is a plastic tissue that adapts in response to many stimuli and demonstrates distinct adaptations to varied exercise modalities (e.g., resistance, endurance, and concurrent or combined exercise). Muscle adaptations to exercise include loading-induced muscle growth (e.g., hypertrophy) as well as metabolic and contractile transformations (e.g., fiber-type transitions and mitochondrial accumulation/remodeling to accommodate the demands placed on the muscle cell).
At the center of adaptations in skeletal muscle are the nuclei- the “brains” of the cell- called myonuclei. Most cell types throughout the body have a single nucleus; however, the long, cylindrical, and voluminous skeletal muscle fibers (myofibers) contain hundreds to thousands of myonuclei. Myofiber multinucleation is the result of the fusion of numerous mononucleated precursor cells during development.9,10 These precursor cells ultimately become muscle stem cells, or satellite cells (SCs), in adult skeletal muscle. The myofiber syncytium is believed to be post-mitotic. Myonuclei do not undergo division, nor is the myofiber thought to divide or “split” at an appreciable level.11,12 Thus, fusion of SCs is required if additional myonuclei or myonuclear replacement is required. The precise cellular and molecular contributions of resident myonuclei versus SC-derived myonuclei in exercise adaptation are incompletely understood, making it an area of ongoing investigation.
Only recently have robust models emerged that allow for the dissection of specific molecular contributions from myonuclei to in vivo mammalian adult skeletal muscle exercise adaptation.13 The purpose of this review is to outline contemporary knowledge on the role of myonuclei, both resident and SC-derived, during exercise adaptation in vivo. Many studies infer myonuclear contributions to exercise adaptation from tissue samples, which is a reasonable approach, but we aim to focus on studies specifically and intentionally examining myonuclei in adult muscle using direct measures. In presenting this overview, we identify specific gaps in knowledge and provide guidance on future directions of research.
A brief primer on heterogeneity of nuclei in skeletal muscle
The skeletal muscle environment contains a diversity of nuclei found in mononuclear cells outside of the multinuclear myofiber. These cells include SCs, fibro/adipogenic progenitors (FAPs), immune cells, endothelial cells, and tenocytes, to name a few. Under resting conditions, myonuclei comprise ∼50%–70% of all nuclei within the muscle tissue.14,15 When murine muscle is subject to acute mechanical overload (a rapid hypertrophic stimulus), the myonuclear proportion can drop to ∼30% of all nuclei.14 This shift in proportion is primarily due to infiltration and proliferation of non-muscle cell types such as fibrogenic and immune cells (e.g., macrophages and neutrophils). The relative proportion of myonuclei at rest, as well as the changes that can occur under dynamic muscle loading conditions highlights the complexity of myonuclear contributions to adaptation. It can be reasonably inferred that certain genes are being expressed by myonuclei in muscle tissue since they are muscle fiber-specific (e.g. myosin heavy chains, skeletal muscle actin, myoglobin, muscle creatine kinase, etc.), but advances in single cell RNA-sequencing technology reveal the diverse influence of mononuclear cell types to the overall gene expression profile of muscle in stressed conditions.16 Furthermore, myonuclei have specificity for maintaining specialized regions of the myofiber; examples include the cell body, neuromuscular junction, and myotendinous junction-associated myonuclei.17,18 Myonuclear subpopulations can complicate the interpretation of myonuclear contributions to exercise. There are clear differences in myonuclear density according to myosin fiber type,19, 20, 21, 22 which points to fiber type-specific differences in myonuclear behavior. The specific influence of SC-acquired myonuclei during exercise adaptation are also poorly understood. Overall, the molecular contributions of myonuclei to exercise adaptation is an area of open inquiry.
The regulation of transcription according to myonuclear number in response to loading
An intuitive task that the myonucleus performs to support exercise adaptations is transcription of muscle-specific genes coding for contractile elements, as well as excitation-contraction coupling, extracellular matrix, metabolism, and ribosomal genes. The latter is especially prevalent since ∼85% of RNA in muscle is ribosomal RNA (rRNA).23 One potential method to increase transcription of protein coding genes and rRNA as adult myofibers adapt to exercise is more myonuclei. It is well-established that myonuclear accretion occurs via fusion of SCs to the myofiber following endurance, resistance, and concurrent exercise, in the presence or absence of myofiber hypertrophy.19,24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37 Irrespective of the cause of myonuclear addition in response to exercise, how new myonuclei contribute to myofiber adaptation at the molecular level can be difficult to discern.38 The reason for a lack of specific evidence is largely technical; it is difficult to track, isolate, and interrogate myonuclei in a syncytial cell in vivo.39 Using different genetically modified mouse models, recent evidence suggests that newly-fused myonuclei contribute specific transcription factors and ribosomal proteins to growing myofibers as a consequence of synergist ablation-induced mechanical overload (SA) or high-volume hypertrophic progressive weighted wheel running (PoWeR).40,41 Apart from these recent studies, it is assumed that increased myonuclear number generally amplifies transcriptional potential, which could support adaptation in myofibers.35,42 Interestingly, on a per-nucleus basis, global transcription rate in the early phases of loading-induced hypertrophy appears lower when myonuclei are added to an adult muscle fiber than when not added.43 Apart from the regenerative roles of SCs in response to highly damaging exercise,38,44 there is minimal evidence regarding the specific myonuclear contributions to adult muscle fiber hypertrophy in the context of exercise.
Myonuclear transcriptional responses to resistance-type exercise in vivo
Following SA mechanical overload of the mouse plantaris, which is a well-documented hypertrophic stimulus, transcription upregulates up to 7-fold in muscle tissue within a few days.43 Up to 14 days after overload, the majority of transcription in muscle during hypertrophic loading is reportedly myonuclear.43 Without myonuclear accretion from SCs, resident myonuclei can upregulate transcription during myofiber hypertrophy, providing evidence of myonuclear transcriptional reserve in growing adult muscle.43 Given most RNA in muscle is rRNA, and rRNA increases dramatically in response to resistance exercise (RE) but not endurance exercise (EE),45 it can be inferred that a large proportion of myonuclear transcription is ribosomal. Ribosome biogenesis is a process thought to be essential for sustained loading-induced muscle growth.46, 47, 48
Global epigenetic profiling of myonuclei after acute SA in mice further points to growth-related transcription by myonuclei.14 Generally, myonuclear DNA hypomethylation occurs after acute short-term loading.14 Specifically, hypomethylation of the promoter region of the oncogene and ribosome biogenesis-related transcription factor Myc occurs with SA, along with promoter sites in numerous muscle growth and autophagy genes.14,45 There is also differential methylation in areas of ribosomal DNA.14,45 It is unclear whether myonuclear methylome changes after an acute or short-term phase of muscle loading (hours to days) are persistent for a long period of time after loading has ceased. RNA-sequencing (RNA-seq) in myonuclei of short-term overloaded muscle corroborates upregulation of Myc at the gene expression level.49 Recent evidence suggests the powerful transcription factor Myc is central to the muscle hypertrophy-associated gene expression program.49 Myonuclear RNA-seq further reveals robustly elevated levels of extracellular matrix (ECM) and Rho-GTPase genes by the myofiber during the early phase of rapid muscle hypertrophy.49 Although myonuclei may account for a large portion of transcription in muscle tissue during hypertrophy, other cell types such as SCs and fibro/adipogenic progenitors also contribute to muscle gene expression in various ways and to varying degrees.49 Using single myonuclear RNA-seq, evidence indicates the presence of SCs may influence myonuclear transcription in response to acute PoWeR exercise independent from fusion to the myofiber.17,50
In addition to murine studies evaluating the acute myonuclear gene expression contribution to myofiber growth, human studies report gene expression in isolated myofibers following a bout of RE and with training. It is assumed most genes detected by this method are transcribed by myonuclei. In young adults, fast-twitch myosin heavy chain (MyHC) 2A fibers are more responsive than MyHC 1 fibers to acute RE at the gene expression level; this included a variety of genes implicated in myofiber hypertrophy.51,52 One gene, the TWEAK receptor (Tnfrsf12a or Fn14), is highly upregulated by acute RE as well as training in pools of MyHC 2A fibers. Young adults are also more responsive than older adults to acute and chronic RE, regardless of myofiber type- in line with the observation that older adults are less responsive to RE training.52, 53, 54 However, caution should be used when interpreting isolated myofiber gene expression. Mononuclear cells such as SCs can adhere to myofibers after mechanical isolation,14 potentially influencing gene expression profiles. To this point, activated muscle stem cells are the cell type most enriched for Fn14 in skeletal muscle.16
With respect to resistance-type training, a few studies evaluated myonuclear epigenetic contributions to adaptation and show responses to chronic exercise may be partly distinct from acute exercise. Eight weeks of PoWeR alters the myonuclear DNA methylation landscape in resident myonuclei (e.g., myonuclei present at the start of training) of the plantaris muscle of young mice.37 Genes in myonuclei with training-induced promoter region hypomethylation correspond with processes linked to protein turnover, mitochondrial biogenesis, and cellular remodeling, such as Wnt and NF-κB signaling. Following PoWeR in mice from 22 to 24 months of age, soleus myonuclear DNA methylation was characterized by global hypomethylation across genomic features including promoters.36 A subset of genes with differential promoter methylation from training also have changes in gene expression corresponding with methylation status. Apart from these studies, there is little high-resolution myonucleus-specific in vivo information available regarding hypertrophic exercise adaptation.
The current evidence collectively indicates that myonuclear transcription contributes to muscle hypertrophy with resistance-type exercise in vivo, and that complimentary processes such as ECM gene expression and Myc induction are regulated in the myofiber. Some changes in myonuclear gene expression are linked to changes in the DNA methylation status of myonuclear DNA. Applying modern technologies such as single myonuclear RNA-seq and ATAC-seq to exercised muscle tissue in vivo will provide more granular insight on myonucleus-autonomous events regulating hypertrophy.
Myonuclear gene expression responses to resistance-type exercise in vitro
In vitro models of RE utilizing electrical pulse stimulation (EPS) have emerged as a method to study muscle adaptation to exercise.55, 56, 57 Such models can provide insight into the specific contributions of myonuclei to hypertrophic exercise since only myogenic cells are used to generate myotubes in culture. In murine-derived C2C12 myotubes, these models reveal a decreased expression of MSTN and increased expression of IGF-1, MGF, and MMP9.55,56 In one model, altered gene expression overlaps significantly between acute myotube stimulation in culture and in vivo RE in human muscle tissues; however, in vitro RE did not mimic the epigenetic response to in vivo RE.56 The dissimilarity in epigenetic modifications between in vitro and in vivo RE may be due to contraction type and mechanical factors (e.g., deformation, maximum tension, and changes in muscle length that occur in vivo), or indicate that epigenetic modifications are differential in vitro versus in vivo, perhaps due to the influence of non-muscle cell types. Hypertrophic stimuli may also modulate translocation of nuclear proteins (e.g., transcription factors). Capsaicin-induced hypertrophy of myotubes increase the migration distance of high molecular weight (55–110 kDa) nuclear proteins derived from a given myonucleus in the syncytium. Uptake of proteins generated by neighboring myonuclei may function as a signal to coordinate transcription during growth, but more work is needed to understand the mechanisms of this process.58
Myonuclear gene expression responses to endurance-type exercise
To our knowledge, investigators have not performed in vivo models of EE in conjunction with myonuclear isolation. As a surrogate, exercise models utilizing EPS in vitro with C2C12 myotubes are used to mimic in vivo models of EE. These models involve varying stimulation frequency, amplitude, and duration used to simulate training in myofibers.57,59, 60, 61, 62 An in vitro approach can provide some insight on myonucleus-specific contributions to EE adaptation without the influence of other cell types. Using these models, upregulated transcription of the MyHC 1 gene, myogenic factors, and genes involved in mitochondrial biogenesis such as PGC-1α occurs in myotubes in vitro.57,59, 60, 61, 62 An ex vivo model of EE in isolated mouse soleus myofibers similarly demonstrates enhanced gene expression of PGC-1α, PPAR-δ, and PDK4 concomitant with hypomethylation of their respective promoters.63 The regulation of these genes in myofibers can drive fiber type transitioning and metabolic changes.64, 65, 66, 67 The gap in knowledge regarding direct measures of myonuclear gene expression in vivo with EE training warrants further attention.
Myonuclear mobility and morphology with exercise
The myonucleus may modulate transcription to promote exercise adaptation by moving within the myofiber, changing shape, and functioning as a mechanosensor.68, 69, 70 Muscle damage often occurs in response to unaccustomed exercise and eccentric contractions.71,72 Emerging evidence suggests following acute contraction-induced muscle damage, myonuclei can move along the myofiber to the site of injury to aid in localized delivery of mRNA and enhance protein synthesis for muscle sarcomere repair.73 Myonuclei are generally positioned at the periphery of the cell along the path of capillaries and will realign themselves as new capillaries form.74 Myonuclear spatial organization may also “optimize” domains without physically disrupting the continuity of the myofibril network.42,70,75,76 When myonuclei move from a more peripheral location to a site of damage after more severe injury, they may relocate to the center of the myofiber and disproportionally contribute to transcriptional activity.77 Following a week of wheel running in mice, bona fide myonuclei that were genetically labeled prior to exercise are found in the center of myofibers.78 Perhaps this myonuclear translocation is associated with movement toward sites of sarcolemmal and/or sarcomere repair to facilitate adaptation to exercise. Recent evidence also suggests that myonuclei are less elongated with different types of exercise training in rodents.78,79 Myonuclear shape change with exercise could be related to their post-training transcriptional status and role as a mechanosensor.69 Some evidence suggests that PGC-1α expression, which is induced by exercise in muscle, can alter myonuclear shape.80 More work is needed to elucidate the function of myonuclear mobility and shape change in the context of exercise adaptation.
Myofiber enriched microRNAs (myomiRs) and their role during exercise
MicroRNAs (miRNAs or miRs) are short, non-coding RNA that affect mRNA stability and translational efficiency, consequently changing protein levels without altering the genetic code. A subclass of miRNAs known as myomiRs are enriched in striated muscle and are sensitive to EE and RE training.81, 82, 83 As myomiRs are expressed almost exclusively in skeletal muscle myofibers, it can be inferred they are produced mostly by myonuclei. MyomiRs appear integral to regulating muscle growth, atrophy, and fiber type switching, but much is yet to be discovered about their functions in muscle.84, 85, 86 The most abundant myomiR in skeletal muscle, miR-1, is a presumptive negative regulator of muscle mass by inhibiting factors in the IGF-1/PI3k/AKT axis, consequently blunting protein synthesis.79,87 MiR-1 also inhibits the gene target of glucose-6-phosphate dehydrogenase (G6pdx), the rate-limiting enzyme in the pentose phosphate pathway, potentially contributing to metabolic reprogramming during muscle loading.88,89 MiR-1 decreases acutely and chronically in response to RE training.77,90, 91, 92, 93, 94 Conversely, acute bouts of EE can increase miR-1.83,95 MiR-206 is another myomiR relevant to exercise adaptations and could facilitate fiber-type transitioning by inhibiting transcriptional repressors of the MyHC 1 gene.82,84,90,96 Worth mentioning, though, miR-206 is also abundant in SCs and subsets of FAPs.97, 98, 99, 100, 101 Therefore, its presence in muscle tissue may not always be attributable to myonuclear transcription.
Myonuclear histone modifications and exercise
Histone modifications, including acetylation, methylation, phosphorylation, and ubiquitination, may epigenetically regulate gene expression.102, 103, 104 Literature regarding exercise-induced histone modifications exclusively in myonuclei is scant, limiting the depth of our commentary. In mice, four weeks of voluntary running increases histone turnover (proxied by incorporation of H2B-GFP into nucleosomes) in conjunction with fewer histones.105 These histone adaptations cause loosening of nucleosomes and may lead to increased gene expression. Following acute SA, histone H3 acetylation, which typically coincides with increased transcription, was greater in myonuclei.43 Further, acute bouts of forced eccentric muscle contractions and downhill treadmill running in mice induced a transient elevation in phosphorylation and acetylation of histone H3 in myonuclei.106 These studies indicate that exercise may modify histones in a manner that allows for modulating accessibility to the DNA sequence and thus gene expression.
Myonuclear “memory” of past training adaptations
In the context of sport performance, “muscle memory” usually refers to rapid re-acquisition of muscular strength or sport skills. The mechanism for this re-acquisition is at the intersection of motor learning, neuromuscular adaptations, and longer-lasting changes to skeletal muscle fibers after prolonged periods of detraining.107 Several lines of evidence direct the molecular explanation for “muscle memory” towards myonuclei. At the cellular level, one proposed mechanism for muscle memory is permanence of myonuclei acquired by SCs during exercise training.108 Recent review papers and cross-talk debates have discussed the plausibility of myonuclear permanence with detraining, atrophy, and aging in great detail.109, 110, 111, 112, 113, 114, 115 In summary, the current evidence suggests that myonuclei gained during exercise training may constitute a muscle memory in the short term (weeks to months), but are likely not permanent over the long term. The maintenance of myonuclei gained during exercise training may therefore not be a definitive explanation for muscle memory. That said, myonuclear loss may also have the prerequisite of significant muscle atrophy (> 30%) and be muscle-type or myofiber-type specific.78,109 Technical issues may also contribute this contested area of inquiry. For example, results from a recent meta-analysis indicate that denervation- the most commonly used model of myofiber atrophy in rodents- causes a significant increase in the number of SCs. Satellite cell behavior and/or abundance may influence the process of maintaining myonuclear number during atrophy.109 It is also imperative to clearly identify the myofiber cell border to ensure non-myonuclear cells are not mis-identified. The complexity of myonuclear identification may exacerbate challenges when quantifying myonuclei via cross section or isolated single fibers. To navigate these challenges, future investigations should use models of genetic myonuclear labeling to ensure exclusively resident myonuclei are quantified.13,78
While data are limited in the current literature, other prospective mechanisms for muscle memory are long-lasting changes to methylation in certain regions of myonuclear DNA, as well as altered miRNA expression. DNA methylation is a dynamic process and, as previously discussed, both EE and RE alter the DNA methylome. One rodent study reported that after chronic PoWeR training and a period of detraining (return to baseline myofiber size), myonuclear DNA partially maintains its differential methylation status months later, particularly in genes involved in muscle hypertrophy.37 Upon a month of retraining, previously trained mice have accelerated muscle hypertrophy. Studies utilizing human muscle tissue corroborate long-term changes to muscle DNA methylation after training.116, 117, 118 There is some reversion of modifications to the DNA methylome to pre-training state after detraining. However, when previously trained individuals retrain, they more rapidly restore modifications to the DNA methylome, along with muscle mass/strength and myofiber size compared to untrained individuals.116,118 A similar pattern is shown with respect to miRNAs. After two months of PoWeR training in mice, miR-1 levels are significantly reduced, which persists after six months of detraining.78 Perhaps myomiR expression is subject to epigenetic regulation as well.
Collectively, the concept of cellular muscle memory has an evidence-based foundation, but the mechanisms of muscle memory are not fully elucidated. Myonuclear permanence over extended periods of time is one possibility. An alternative explanation is that epigenetic modifications to myonuclear DNA allow for genes involved in hypertrophic adaptation to be more readily expressed once training resumes, thereby facilitating more rapid adaptation.
Summary
Through recently developed technologies such as inducible myonuclear labeling and single myonuclear RNA-seq (smnRNA-seq), researchers can overcome the barrier of analyzing heterogenous nuclear populations in skeletal muscle. These advances can allow for the dissection of specific molecular contributions of resident and SC-derived myonuclei to exercise adaptation in vivo. Prior to the utilization of these modern technologies, it could be reasonably inferred that transcription is augmented in response to exercise partly due to fusion of SCs to the myofiber during both EE and RE training. Enhanced transcription may even occur in the absence of hypertrophy; some evidence suggests individual myonuclei in adult muscle can upregulate transcription several-fold. Alterations in myonuclear gene expression driving muscle hypertrophy with resistance-type exercise in vivo are linked to changes in DNA methylation status and myomiR expression, particularly in genes important for growth, autophagy, extracellular matrix remodeling, and ribosome biogenesis. Less is known about myonucleus-specific gene expression changes following endurance-type exercise in vivo. In vitro models show elevated mRNA expression of genes involved in fast-to-slow fiber type switching, myogenic factors, and mitochondrial biogenesis. Transcription profiles in resident and SC-derived myonuclei may be distinct and appear partly regulated by DNA methylation. Myonuclei may also have a muscle memory of past training adaptations. We posit this memory is more likely due to epigenetic modifications to the DNA methylome and/or changes in myomiR expression, and less likely myonuclear permanence. Precisely what drives epigenetic modifications in myonuclei is poorly understood. Taken together, the current evidence provides a preliminary understanding of the myonuclear functions that support exercise adaptations (Fig. 1), but there are still numerous gaps in knowledge.
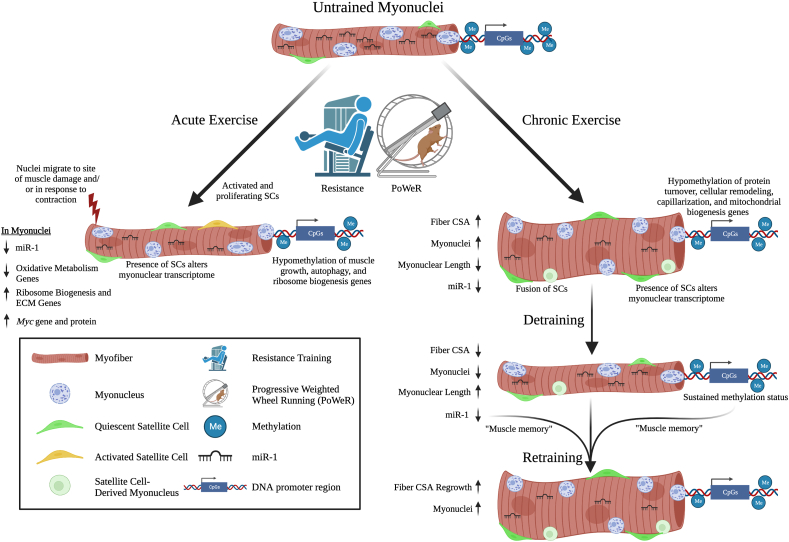
Fig. 1.
With acute exercise, myonuclei can migrate along the myofiber to a site of damage and aid in localized mRNA delivery to facilitate muscle sarcomere repair. Myonuclear movement is likely of particular relevance with unaccustomed exercise featuring an eccentric (damaging) component, but some evidence also suggests that myonuclei can move as a consequence of contraction. Satellite cells activate and may proliferate in response to several acute exercise-induced signals. The presence of satellite cells can also influence the myonuclear transcriptome independent from fusion, as revealed by satellite cell loss-of-function studies. Epigenetic modifications that occur in myonuclear DNA after exercise include hypomethylation in promoter regions of genes involved in muscle growth, autophagy, and ribosomal biogenesis, along with lower expression of miR-1. Epigenetic modifications in muscle fibers may result in transiently reduced expression of oxidative metabolism genes, as well as increased expression of extracellular matrix and ribosomal biogenesis genes. With chronic exercise training, myofiber size and myonuclear density increase due to fusion of satellite cells, and a decrease in myonuclear length. The myonuclear transcriptome may be altered by the presence of satellite cells and epigenetic modifications to myonuclear DNA such as hypomethylation of genes involved in protein turnover, cellular remodeling, capillarization, and mitochondrial biogenesis, as well as chronically reduced expression of miR-1. Epigenetic modifications are sustained with detraining and may function as a “muscle memory” to potentiate a rapid re-acquisition of training adaptations upon retraining. Figure was generated using BioRender.
Future directions in studying myonuclei
New models of murine exercise training are rapidly developing.119,120 Combining murine exercise with pre-clinical models of genetic myonuclear labeling will provide a robust platform for studying myonuclear adaptations to different forms of exercise.13,39 Developing new methods to track SC-derived myonuclei in vivo will further enhance these efforts. Directing effort toward understanding how SC-derived myonuclei contribute to specific fiber types could also be worthwhile since myonuclear addition with training can differ according to MyHC isoform.25 In humans, the utilization of an antibody to specifically isolate myonuclei will enable high-resolution analysis of myonuclear adaptations to exercise,121 although recent evidence suggests a more specific antibody may be required, specifically during times of muscle stress.122 High-resolution sequencing of RNA, miRNA, chromatin accessibility, DNA methylation, and other molecular layers at single nucleus resolution in mice and humans will provide molecular maps of exercise adaptation across age ranges and sexes, and further reveal subpopulations and heterogeneity among myonuclei in dynamic conditions.
Submission statement
All authors have read and approve of the contents of the manuscript. While the manuscript is under review for this journal the manuscript will not be submitted elsewhere for review and publication.
Authors' contributions
Pieter J. Koopmans drafted the manuscript and created the figure. Kevin A. Murach and Kevin A. Zwetsloot provided critical feedback, edited, and revised the manuscript. All authors approve of the final version of the manuscript.
Conflict of interest
The authors have no conflicts to declare.
References
- 1.Frontera WR, Ochala J. Skeletal Muscle: a brief review of structure and function. Calcif Tissue Int. 2015;96(3):183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 2.Demontis F, Piccirillo R, Goldberg AL, Perrimon N. The influence of skeletal muscle on systemic aging and lifespan. Aging Cell. 2013;12(6):943–949. doi: 10.1111/acel.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiao J, Demontis F. Skeletal muscle autophagy and its role in sarcopenia and organismal aging. Curr Opin Pharmacol. 2017;34:1–6. doi: 10.1016/j.coph.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Pérez-Schindler J, Handschin C. In: Nutrition and Skeletal Muscle. Walrand S, editor. Academic Press; 2019. Physiological regulation of skeletal muscle mass: resistance exercise-mediated muscle hypertrophy; pp. 139–150. [DOI] [Google Scholar]
- 5.McLeod M, Breen L, Hamilton DL, Philp A. Live strong and prosper: the importance of skeletal muscle strength for healthy ageing. Biogerontology. 2016;17(3):497–510. doi: 10.1007/s10522-015-9631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartee GD, Hepple RT, Bamman MM, Zierath JR. Exercise promotes healthy aging of skeletal muscle. Cell Metabol. 2016;23(6):1034–1047. doi: 10.1016/j.cmet.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Booth FW, Roberts CK, Laye MJ. In: Comprehensive Physiology. first ed. Terjung R., editor. Wiley; 2012. Lack of exercise is a major cause of chronic diseases; pp. 1143–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson E, Durstine JL. Physical activity, exercise, and chronic diseases: a brief review. Sports Med Health Sci. 2019;1(1):3–10. doi: 10.1016/j.smhs.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert SF. sixth ed. 2000. Myogenesis: The Development of Muscle. Dev Biol.https://www.ncbi.nlm.nih.gov/books/NBK10006/ Published online. [Google Scholar]
- 10.Bruusgaard JC, Liestøl K, Ekmark M, Kollstad K, Gundersen K. Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in Vivo. J Physiol. 2003;551(2):467–478. doi: 10.1113/jphysiol.2003.045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partridge TA. Cells that participate in regeneration of skeletal muscle. Gene Ther. 2002;9(11):752–753. doi: 10.1038/sj.gt.3301764. [DOI] [PubMed] [Google Scholar]
- 12.Chal J, Pourquié O. Making muscle: skeletal myogenesis in vivo and in vitro. Dev Camb Engl. 2017;144(12):2104–2122. doi: 10.1242/dev.151035. [DOI] [PubMed] [Google Scholar]
- 13.Iwata M, Englund DA, Wen Y, et al. A novel tetracycline-responsive transgenic mouse strain for skeletal muscle-specific gene expression. Skeletal Muscle. 2018;8(1):33. doi: 10.1186/s13395-018-0181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Von Walden F, Rea M, Mobley CB, et al. The myonuclear DNA methylome in response to an acute hypertrophic stimulus. Epigenetics. 2020;15(11):1151–1162. doi: 10.1080/15592294.2020.1755581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dos Santos M, Backer S, Saintpierre B, et al. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat Commun. 2020;11(1):5102. doi: 10.1038/s41467-020-18789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKellar DW, Walter LD, Song LT, et al. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration. Commun Biol. 2021;4(1):1–12. doi: 10.1038/s42003-021-02810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen Y, Englund DA, Peck BD, Murach KA, McCarthy JJ, Peterson CA. Myonuclear transcriptional dynamics in response to exercise following satellite cell depletion. iScience. 2021;24(8):102838. doi: 10.1016/j.isci.2021.102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M, Franke V, Brandt B, et al. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat Commun. 2020;11(1):6375. doi: 10.1038/s41467-020-20064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moro T, Brightwell CR, Volpi E, Rasmussen BB, Fry CS. Resistance exercise training promotes fiber type-specific myonuclear adaptations in older adults. J Appl Physiol. 2020;128(4):795–804. doi: 10.1152/japplphysiol.00723.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng BS, Kasper CE, Edgerton VR. Cytoplasm-to-myonucleus ratios and succinate dehydrogenase activities in adult rat slow and fast muscle fibers. Cell Tissue Res. 1994;275(1):39–49. doi: 10.1007/BF00305374. [DOI] [PubMed] [Google Scholar]
- 21.Burleigh IG. Observations on the number of nuclei within the fibres of some red and white muscles. J Cell Sci. 1977;23(1):269–284. doi: 10.1242/jcs.23.1.269. [DOI] [PubMed] [Google Scholar]
- 22.Davey D, Wong S. Morphometric analysis of rat extensor digitorum longus and soleus muscles. Aust J Exp Biol Med Sci. 1980;58(3):213–230. doi: 10.1038/icb.1980.22. [DOI] [PubMed] [Google Scholar]
- 23.Zak R, Rabinowitz M, Platt C. Ribonucleic acids associated with myofibrils. Biochemistry. 1967;6(8):2493–2500. doi: 10.1021/bi00860a028. [DOI] [PubMed] [Google Scholar]
- 24.Frese S, Ruebner M, Suhr F, et al. Long-term Endurance Exercise in Humans Stimulates Cell Fusion of Myoblasts along with Fusogenic Endogenous Retroviral Genes in Vivo. PLoS One. 2015;10(7):e0132099. doi: 10.1371/journal.pone.0132099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fry CS, Noehren B, Mula J, et al. Fibre type-specific satellite cell response to aerobic training in sedentary adults: fibre type satellite cell content increases with aerobic training. J Physiol. 2014;592(12):2625–2635. doi: 10.1113/jphysiol.2014.271288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenzie AI, D’Lugos AC, Saunders MJ, Gworek KD, Luden ND. Fiber type-specific satellite cell content in cyclists following heavy training with carbohydrate and carbohydrate-protein supplementation. Front Physiol. 2016;7:550. doi: 10.3389/fphys.2016.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goh Q, Song T, Petrany MJ, et al. Myonuclear accretion is a determinant of exercise-induced remodeling in skeletal muscle. Elife. 2019;8:e44876. doi: 10.7554/eLife.44876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurosaka M, Naito H, Ogura Y, Kojima A, Goto K, Katamoto S. Effects of voluntary wheel running on satellite cells in the rat plantaris muscle. J Sports Sci Med. 2009;8(1):51–57. [PMC free article] [PubMed] [Google Scholar]
- 29.Smith HK, Maxwell L, Rodgers CD, McKee NH, Plyley MJ. Exercise-enhanced satellite cell proliferation and new myonuclear accretion in rat skeletal muscle. J Appl Physiol. 2001;90(4):1407–1414. doi: 10.1152/jappl.2001.90.4.1407. [DOI] [PubMed] [Google Scholar]
- 30.Bjørnsen T, Wernbom M, Løvstad A, et al. Delayed myonuclear addition, myofiber hypertrophy, and increases in strength with high-frequency low-load blood flow restricted training to volitional failure. J Appl Physiol. 2019;126(3):578–592. doi: 10.1152/japplphysiol.00397.2018. [DOI] [PubMed] [Google Scholar]
- 31.Mobley CB, Holland AM, Kephart WC, et al. Progressive resistance-loaded voluntary wheel running increases hypertrophy and differentially affects muscle protein synthesis, ribosome biogenesis, and proteolytic markers in rat muscle. J Anim Physiol Anim Nutr. 2018;102(1):317–329. doi: 10.1111/jpn.12691. [DOI] [PubMed] [Google Scholar]
- 32.Snijders T, Smeets JSJ, van Kranenburg J, Kies AK, van Loon LJC, Verdijk LB. Changes in myonuclear domain size do not precede muscle hypertrophy during prolonged resistance-type exercise training. Acta Physiol. 2016;216(2):231–239. doi: 10.1111/apha.12609. [DOI] [PubMed] [Google Scholar]
- 33.Petrella JK, Kim J, Mayhew DL, Cross JM, Bamman MM. Potent myofiber hypertrophy during resistance training in humans is associated with satellite cell-mediated myonuclear addition: a cluster analysis. J Appl Physiol. 2008;104(6):1736–1742. doi: 10.1152/japplphysiol.01215.2007. [DOI] [PubMed] [Google Scholar]
- 34.Snijders T, Holwerda AM, Loon LJC, Verdijk LB. Myonuclear content and domain size in small versus larger muscle fibres in response to 12 weeks of resistance exercise rraining in older adults. Acta Physiol. 2021;231(4):e13599 doi: 10.1111/apha.13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruusgaard JC, Johansen IB, Egner IM, Rana ZA, Gundersen K. Myonuclei acquired by overload exercise precede hypertrophy and are not lost on detraining. Proc Natl Acad Sci USA. 2010;107(34):15111–15116. doi: 10.1073/pnas.0913935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dungan CM, Brightwell C, Wen Y, et al. Muscle-specific cellular and molecular adaptations to late-life voluntary concurrent exercise. Function. 2022;3(4):zqac027. doi: 10.1093/function/zqac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen Y, Dungan CM, Mobley CB, Valentino T, von Walden F, Murach KA. Nucleus type-specific DNA methylomics reveals epigenetic “memory” of prior adaptation in skeletal muscle. Function. 2021;2(5):zqab038. doi: 10.1093/function/zqab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murach KA, Fry CS, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Fusion and beyond: satellite cell contributions to loading-induced skeletal muscle adaptation. Faseb J. 2021;35(10):e21893. doi: 10.1096/fj.202101096R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masschelein E, D’Hulst G, Zvick J, et al. Exercise promotes satellite cell contribution to myofibers in a load-dependent manner. Skeletal Muscle. 2020;10(1):21. doi: 10.1186/s13395-020-00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murach KA, Peck BD, Policastro RA, et al. Early satellite cell communication creates a permissive environment for long-term muscle growth. iScience. 2021;24(4):102372. doi: 10.1016/j.isci.2021.102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murach KA, Dungan CM, von Walden F, Wen Y. Epigenetic evidence for distinct contributions of resident and acquired myonuclei during long-term exercise adaptation using timed in vivo myonuclear labeling. Am J Physiol Cell Physiol. 2022;322(1):C86–C93. doi: 10.1152/ajpcell.00358.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansson KA, Solbrå AV, Gundersen K, Bruusgaard JC. Computational assessment of transport distances in living skeletal muscle fibers studied in situ. Biophys J. 2020;119(11):2166–2178. doi: 10.1016/j.bpj.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirby TJ, Patel RM, McClintock TS, Dupont-Versteegden EE, Peterson CA, McCarthy JJ. Myonuclear transcription is responsive to mechanical load and DNA content but uncoupled from cell size during hypertrophy. Marshall W. Mol Biol Cell. 2016;27(5):788–798. doi: 10.1091/mbc.E15-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackey AL, Kjaer M. The breaking and making of healthy adult human skeletal muscle in vivo. Skeletal Muscle. 2017;7(1):24. doi: 10.1186/s13395-017-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figueiredo VC, Wen Y, Alkner B, et al. Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. J Physiol. 2021;599(13):3363–3384. doi: 10.1113/JP281244. [DOI] [PubMed] [Google Scholar]
- 46.Wen Y, Alimov AP, McCarthy JJ. Ribosome biogenesis is necessary for skeletal muscle hypertrophy. Exerc Sport Sci Rev. 2016;44(3):110–115. doi: 10.1249/JES.0000000000000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Figueiredo VC, McCarthy JJ. Regulation of ribosome biogenesis in skeletal muscle hypertrophy. Physiology. 2019;34(1):30–42. doi: 10.1152/physiol.00034.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Walden F. Ribosome biogenesis in skeletal muscle: coordination of transcription and translation. J Appl Physiol. 2019;127(2):591–598. doi: 10.1152/japplphysiol.00963.2018. [DOI] [PubMed] [Google Scholar]
- 49.Murach KA, Liu Z, Jude B, et al. Multi-transcriptome analysis following an acute skeletal muscle growth stimulus yields tools for discerning global and MYC regulatory networks. J Biol Chem. 2022;298(11):102515. doi: 10.1016/j.jbc.2022.102515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Englund DA, Figueiredo VC, Dungan CM, et al. Satellite cell depletion disrupts transcriptional coordination and muscle adaptation to exercise. Function. 2021;2(1):zqaa033. doi: 10.1093/function/zqaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Myogenic gene expression at rest and after a bout of resistance exercise in young (18–30 yr) and old (80–89 yr) women. J Appl Physiol. 2006;101(1):53–59. doi: 10.1152/japplphysiol.01616.2005. [DOI] [PubMed] [Google Scholar]
- 52.Raue U, Trappe TA, Estrem ST, et al. Transcriptome signature of resistance exercise adaptations: mixed muscle and fiber type specific profiles in young and old adults. J Appl Physiol. 2012;112(10):1625–1636. doi: 10.1152/japplphysiol.00435.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greig CA, Gray C, Rankin D, et al. Blunting of adaptive responses to resistance exercise training in women over 75y. Exp Gerontol. 2011;46(11):884–890. doi: 10.1016/j.exger.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Welle S, Totterman S, Thornton C. Effect of age on muscle hypertrophy induced by resistance training. J Gerontol A Biol Sci Med Sci. 1996;51A(6):M270–M275. doi: 10.1093/gerona/51A.6.M270. [DOI] [PubMed] [Google Scholar]
- 55.Tarum J, Folkesson M, Atherton PJ, Kadi F. Electrical pulse stimulation: an in vitro exercise model for the induction of human skeletal muscle cell hypertrophy. A proof-of-concept study. Exp Physiol. 2017;102(11):1405–1413. doi: 10.1113/EP086581. [DOI] [PubMed] [Google Scholar]
- 56.Turner DC, Gorski PP, Seaborne RA, et al. Mechanical loading of bioengineered skeletal muscle in vitro recapitulates gene expression signatures of resistance exercise in vivo. J Cell Physiol. 2021;236(9):6534–6547. doi: 10.1002/jcp.30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nikolić N, Görgens SW, Thoresen GH, Aas V, Eckel J, Eckardt K. Electrical pulse stimulation of cultured skeletal muscle cells as a model for in vitro exercise – possibilities and limitations. Acta Physiol. 2017;220(3):310–331. doi: 10.1111/apha.12830. [DOI] [PubMed] [Google Scholar]
- 58.Taylor-Weiner H, Grigsby CL, Ferreira DMS, et al. Modeling the transport of nuclear proteins along single skeletal muscle cells. Proc Natl Acad Sci USA. 2020;117(6):2978–2986. doi: 10.1073/pnas.1919600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Son YH, Lee SM, Lee SH, et al. Comparative molecular analysis of endurance exercise in vivo with electrically stimulated in vitro myotube contraction. J Appl Physiol Bethesda Md 1985. 2019;127(6):1742–1753. doi: 10.1152/japplphysiol.00091.2019. [DOI] [PubMed] [Google Scholar]
- 60.Scheler M, Irmler M, Lehr S, et al. Cytokine response of primary human myotubes in an in vitro exercise model. Am J Physiol Cell Physiol. 2013;305(8):C877–C886. doi: 10.1152/ajpcell.00043.2013. [DOI] [PubMed] [Google Scholar]
- 61.Kurdiova T, Balaz M, Mayer A, et al. Exercise-mimicking treatment fails to increase FNDC5 mRNA & irisin secretion in primary human myotubes. Peptides. 2014;56:1–7. doi: 10.1016/j.peptides.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 62.Naumann K, Pette D. Effects of chronic stimulation with different impulse patterns on the expression of myosin isoforms in rat myotube cultures. Differentiation. 1994;55(3):203–211. doi: 10.1046/j.1432-0436.1994.5530203.x. [DOI] [PubMed] [Google Scholar]
- 63.Barrès R, Yan J, Egan B, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metabol. 2012;15(3):405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 64.Koves TR, Li P, An J, et al. Peroxisome proliferator-activated receptor-γ co-activator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280(39):33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- 65.Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 66.Russell AP, Feilchenfeldt J, Schreiber S, et al. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes. 2003;52(12):2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- 67.Holness MJ, Kraus A, Harris RA, Sugden MC. Targeted upregulation of pyruvate dehydrogenase kinase (PDK)-4 in slow-twitch skeletal muscle underlies the stable modification of the regulatory characteristics of PDK induced by high-fat feeding. Diabetes. 2000;49(5):775–781. doi: 10.2337/diabetes.49.5.775. [DOI] [PubMed] [Google Scholar]
- 68.Azevedo M, Baylies MK. Getting into position: nuclear movement in muscle cells. Trends Cell Biol. 2020;30(4):303–316. doi: 10.1016/j.tcb.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol. 2018;20(4):373–381. doi: 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Folker ES, Baylies MK. Nuclear positioning in muscle development and disease. Front Physiol. 2013;4:363. doi: 10.3389/fphys.2013.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Proske U, Morgan DL. Muscle damage from eccentric exercise: mechanism, mechanical signs, adaptation and clinical applications. J Physiol. 2001;537(Pt 2):333–345. doi: 10.1111/j.1469-7793.2001.00333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murach KA, Mobley CB, Zdunek CJ, et al. Muscle memory: myonuclear accretion, maintenance, morphology, and miRNA levels with training and detraining in adult mice. J Cachexia Sarcopenia Muscle. 2020;11(6):1705–1722. doi: 10.1002/jcsm.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roman W, Pinheiro H, Pimentel MR, et al. Muscle repair after physiological damage relies on nuclear migration for cellular reconstruction. Science. 2021;374(6565):355–359. doi: 10.1126/science.abe5620. [DOI] [PubMed] [Google Scholar]
- 74.Ralston E, Lu Z, Biscocho N, et al. Blood vessels and desmin control the positioning of nuclei in skeletal muscle fibers. J Cell Physiol. 2006;209(3):874–882. doi: 10.1002/jcp.20780. [DOI] [PubMed] [Google Scholar]
- 75.Glancy B, Hsu LY, Dao L, et al. In vivo microscopy reveals extensive embedding of capillaries within the sarcolemma of skeletal muscle fibers. Microcirculation. 2014;21(2):131–147. doi: 10.1111/micc.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Willingham T.B., Ajayi P.T., Glancy B. Subcellular specialization of mitochondrial form and function in skeletal muscle cells. Front Cell Dev Biol. 2021;9:757305. doi: 10.3389/fcell.2021.757305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buckley KH, Nestor-Kalinoski AL, Pizza FX. Positional context of myonuclear transcription during injury-induced muscle regeneration. Front Physiol. 2022;13:845504. doi: 10.3389/fphys.2022.845504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murach KA, Mobley CB, Zdunek CJ, et al. Muscle memory: myonuclear accretion, maintenance, morphology, and miRNA levels with training and detraining in adult mice. J Cachexia Sarcopenia Muscle. 2020;11(6):1705–1722. doi: 10.1002/jcsm.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rader EP, Baker BA. Elevated muscle mass accompanied by transcriptional and nuclear alterations several months following cessation of resistance-type training in rats. Physiol Rep. 2022;10(20):e15476. doi: 10.14814/phy2.15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ross JA, Pearson A, Levy Y, Cardel B, Handschin C, Ochala J. Exploring the role of PGC-1α in defining nuclear organisation in skeletal muscle fibres. J Cell Physiol. 2017;232(6):1270–1274. doi: 10.1002/jcp.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by micrornas. Annu Rev Biochem. 2010;79(1):351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 82.Mccarthy J. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta BBA - Gene Regul Mech. 2008;1779(11):682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nielsen S, Scheele C, Yfanti C, et al. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle: muscle specific microRNAs and exercise. J Physiol. 2010;588(20):4029–4037. doi: 10.1113/jphysiol.2010.189860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giagnorio E, Malacarne C, Mantegazza R, Bonanno S, Marcuzzo S. MyomiRs and their multifaceted regulatory roles in muscle homeostasis and amyotrophic lateral sclerosis. J Cell Sci. 2021;134(12):jcs258349. doi: 10.1242/jcs.258349. [DOI] [PubMed] [Google Scholar]
- 85.Kovanda A, Režen T, Rogelj B. MicroRNA in skeletal muscle development, growth, atrophy, and disease: functions of miRNA in skeletal muscle. Wiley Interdiscip Rev RNA. 2014;5(4):509–525. doi: 10.1002/wrna.1227. [DOI] [PubMed] [Google Scholar]
- 86.Horak M, Novak J, Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev Biol. 2016;410(1):1–13. doi: 10.1016/j.ydbio.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 87.Elia L, Contu R, Quintavalle M, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120(23):2377–2385. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh A, Happel C, Manna SK, et al. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest. 2013;123(7):2921–2934. doi: 10.1172/JCI66353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Valentino T, Figueiredo VC, Mobley CB, McCarthy JJ, Vechetti IJ., Jr. Evidence of myomiR regulation of the pentose phosphate pathway during mechanical load-induced hypertrophy. Phys Rep. 2021;9(23):e15137. doi: 10.14814/phy2.15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102(1):306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 91.Mueller M, Breil FA, Lurman G, et al. Different molecular and structural adaptations with eccentric and conventional strength training in elderly men and women. Gerontology. 2011;57(6):528–538. doi: 10.1159/000323267. [DOI] [PubMed] [Google Scholar]
- 92.Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. 2008;295(6):E1333–E1340. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vechetti IJ, Valentino T, Mobley CB, McCarthy JJ. The role of extracellular vesicles in skeletal muscle and systematic adaptation to exercise. J Physiol. 2021;599(3):845–861. doi: 10.1113/JP278929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. J Appl Physiol. 2013;115(7):1065–1074. doi: 10.1152/japplphysiol.00611.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Russell AP, Lamon S, Boon H, et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J Physiol. 2013;591(18):4637–4653. doi: 10.1113/jphysiol.2013.255695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol. 2007;293(1):C451–C457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 97.Murach KA, Peck BD, Policastro RA, et al. Early satellite cell communication creates a permissive environment for long-term muscle growth. iScience. 2021;24(4):102372. doi: 10.1016/j.isci.2021.102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sandonà M, Consalvi S, Tucciarone L, et al. HDAC inhibitors tune miRNAs in extracellular vesicles of dystrophic muscle-resident mesenchymal cells. EMBO Rep. 2020;21(9):e50863. doi: 10.15252/embr.202050863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fry CS, Kirby TJ, Kosmac K, McCarthy JJ, Peterson CA. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell. 2017;20(1):56–69. doi: 10.1016/j.stem.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Murach KA, Vechetti IJ, Jr., Van Pelt DW, et al. Fusion-independent satellite cell communication to muscle fibers during load-induced hypertrophy. Function. 2020;1(1):zqaa009. doi: 10.1093/function/zqaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wosczyna MN, Perez Carbajal EE, Wagner MW, et al. Targeting microRNA-mediated gene repression limits adipogenic conversion of skeletal muscle mesenchymal stromal cells. Cell Stem Cell. 2021;28(7):1323–1334. doi: 10.1016/j.stem.2021.04.008. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. 2010;105(1):4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 103.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123(19):2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alaskhar Alhamwe B, Khalaila R, Wolf J, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. 2018;14(1):39. doi: 10.1186/s13223-018-0259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ohsawa I, Kawano F. Chronic exercise training activates histone turnover in mouse skeletal muscle fibers. Faseb J. 2021;35(4):e21453. doi: 10.1096/fj.202002027RR. [DOI] [PubMed] [Google Scholar]
- 106.Solagna F, Nogara L, Dyar KA, et al. Exercise-dependent increases in protein synthesis are accompanied by chromatin modifications and increased MRTF-SRF signalling. Acta Physiol. 2020;230(1):e13496. doi: 10.1111/apha.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Staron RS, Leonardi MJ, Karapondo DL, et al. Strength and skeletal muscle adaptations in heavy-resistance-trained women after detraining and retraining. J Appl Physiol. 1991;70(2):631–640. doi: 10.1152/jappl.1991.70.2.631. [DOI] [PubMed] [Google Scholar]
- 108.Gundersen K. Muscle memory and a new cellular model for muscle atrophy and hypertrophy. J Exp Biol. 2016;219(2):235–242. doi: 10.1242/jeb.124495. [DOI] [PubMed] [Google Scholar]
- 109.Rahmati M, McCarthy JJ, Malakoutinia F. Myonuclear permanence in skeletal muscle memory: a systematic review and meta-analysis of human and animal studies. J Cachexia Sarcopenia Muscle. 2022;13:2276-2297. https://doi.org/10.1002/jcsm.13043. [DOI] [PMC free article] [PubMed]
- 110.Snijders T, Aussieker T, Holwerda A, Parise G, Loon LJC, Verdijk LB. The concept of skeletal muscle memory: evidence from animal and human studies. Acta Physiol. 2020;229(3):e13465. doi: 10.1111/apha.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schwartz LM. Skeletal muscles do not undergo apoptosis during either atrophy or programmed cell death-revisiting the myonuclear domain hypothesis. Front Physiol. 2019;9:1887. doi: 10.3389/fphys.2018.01887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schwartz LM, Gundersen K. Cross talk opposing view: myonuclei do not undergo apoptosis during skeletal muscle atrophy. J Physiol. 2022;600(9):2081–2084. doi: 10.1113/JP282381. [DOI] [PubMed] [Google Scholar]
- 113.Schwartz L, Gundersen K. Cross talk rebuttal: schwartz and gundersen. J Physiol. 2022;600(9):2087–2088. doi: 10.1113/JP283001. [DOI] [PubMed] [Google Scholar]
- 114.Kirby TJ, Dupont-Versteegden EE. Cross talk proposal: myonuclei are lost with ageing and atrophy. J Physiol. 2022;600(9):2077–2080. doi: 10.1113/JP282380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kirby TJ, Dupont-Versteegden EE. Cross talk rebuttal: kirby and dupont-versteegden. J Physiol. 2022;600(9):2085–2086. doi: 10.1113/JP283000. [DOI] [PubMed] [Google Scholar]
- 116.Blocquiaux S, Gorski T, Van Roie E, et al. The effect of resistance training, detraining and retraining on muscle strength and power, myofibre size, satellite cells and myonuclei in older men. Exp Gerontol. 2020;133:110860. doi: 10.1016/j.exger.2020.110860. [DOI] [PubMed] [Google Scholar]
- 117.Seaborne RA, Strauss J, Cocks M, et al. Human skeletal muscle possesses an epigenetic memory of hypertrophy. Sci Rep. 2018;8(1):1898. doi: 10.1038/s41598-018-20287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Seaborne RA, Strauss J, Cocks M, et al. Methylome of human skeletal muscle after acute & chronic resistance exercise training. Detrain Retrain. Sci Data. 2018;5(1):180213. doi: 10.1038/sdata.2018.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhu WG, Hibbert JE, Lin KH, et al. Weight pulling: a novel mouse model of human progressive resistance exercise. Cells. 2021;10(9):2459. doi: 10.3390/cells10092459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Murach KA, McCarthy JJ, Peterson CA, Dungan CM. Making mice mighty: recent advances in translational models of load-induced muscle hypertrophy. J Appl Physiol. 2020;129(3):516–521. doi: 10.1152/japplphysiol.00319.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Winje IM, Bengtsen M, Eftestøl E, Juvkam I, Bruusgaard JC, Gundersen K. Specific labelling of myonuclei by an antibody against pericentriolar material 1 on skeletal muscle tissue sections. Acta Physiol. 2018;223(4):e13034. doi: 10.1111/apha.13034. [DOI] [PubMed] [Google Scholar]
- 122.Viggars MR, Owens DJ, Stewart C, Coirault C, Mackey AL, Jarvis JC. PCM1 labelling reveals myonuclear and nuclear dynamics in skeletal muscle across species. Am J Physiol Cell Physiol. 2022:2022. doi: 10.1152/ajpcell.00285.02022. [DOI] [PubMed] [Google Scholar]