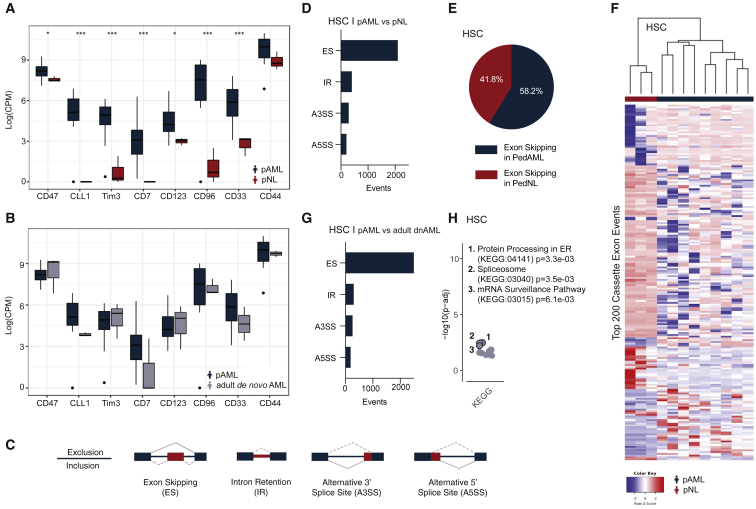
Figure 3.
Whole-transcriptome detection of splicing deregulation in pAML compared with pediatric non-leukemic (pNL) hematopoietic stem cells
(A and B) Transcript expression levels (log counts per million; CPM) of LSC-associated phenotypic markers in pAML (n = 9) versus pNL (n = 3; A) and adult dnAML (n = 3; B) derived hematopoietic stem cells (HSCs; moderated t statistic with Benjamini-Hochberg correction method for multiple testing; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005).
(C) Schematic of alternative splicing event detection.
(D) Quantification of differential splicing events, including exon skipping (ES), intron retention (IR), alternative 3′ splice site usage (A3SS), and alternative 5′ splice site usage (A5SS) in pAML HSCs compared with pNL HSCs using multivariate analysis of transcript splicing (rMATS) bioinformatics analysis.
(E) Percentage of exon skipping in pAML HSCs compared with pNL HSCs.
(F) Heatmap of top 200 distinct splice isoforms in pAML HSCs versus pNL HSCs.
(G) Quantification of differential splicing events (ES, IR, A3SS, A5SS) in pAML HSCs versus adult dnAML HSCs.
(H) Functional enrichment analysis of pAML HSCs versus adult dnAML HSCs. See also Figure S3.