Figure 4.
Lentiviral splicing reporter and RNA sequencing-based detection of splicing alterations in pAML
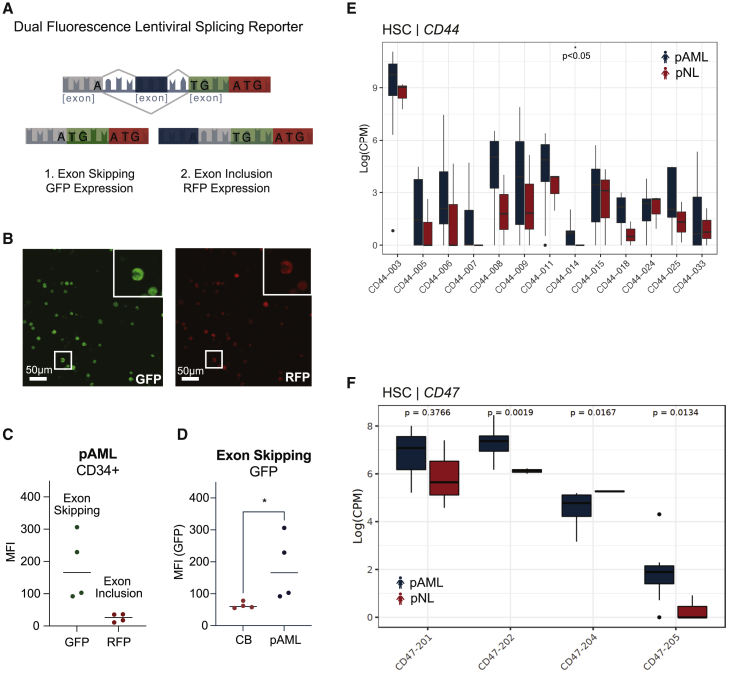
(A) Schematic of lentiviral dual-fluorescence MAPT splicing reporter.
(B) Representative image of Kasumi-1, a pAML cell line, stably transduced with the pCDH-EF1-IRES-Puro MAPT splicing reporter lentiviral vector. Left: confocal fluorescence microscopic image of lentiviral splicing reporter GFP expression (scale bar, 50 μm). Right: confocal fluorescence microscopic image of lentiviral splicing reporter RFP expression (scale bar, 50 μm).
(C) FACS analysis of GFP and RFP mean fluorescence intensity (MFI) in the live cell population of CD34+ selected pAML patient samples (n = 4 biological replicates).
(D) FACS analysis of GFP MFI in the live cell population of CD34+ selected pAML samples versus CD34+ cord blood (CB) samples (Student’s t test, p = 0.029; n = 4 biological replicates).
(E) Whole-transcriptome (RNA-seq)-based quantification of CD44 splice isoforms in pAML (n = 9, blue) versus pNL (n = 3; red) HSCs (Student's t test, p < 0.05).
(F) RNA-seq-based quantification of CD47 splice isoform expression in pAML (blue) versus pNL (red) HSCs (Student's t test, CD47-201 p = 0.3766; CD47-202 p = 0.0019; CD47-204 p = 0.0167; CD47-205 p = 0.0134). See also Figure S4.