Figure 5.
RBFOX2 downregulation and reversion to an embryonic stem cell splicing program in pAML
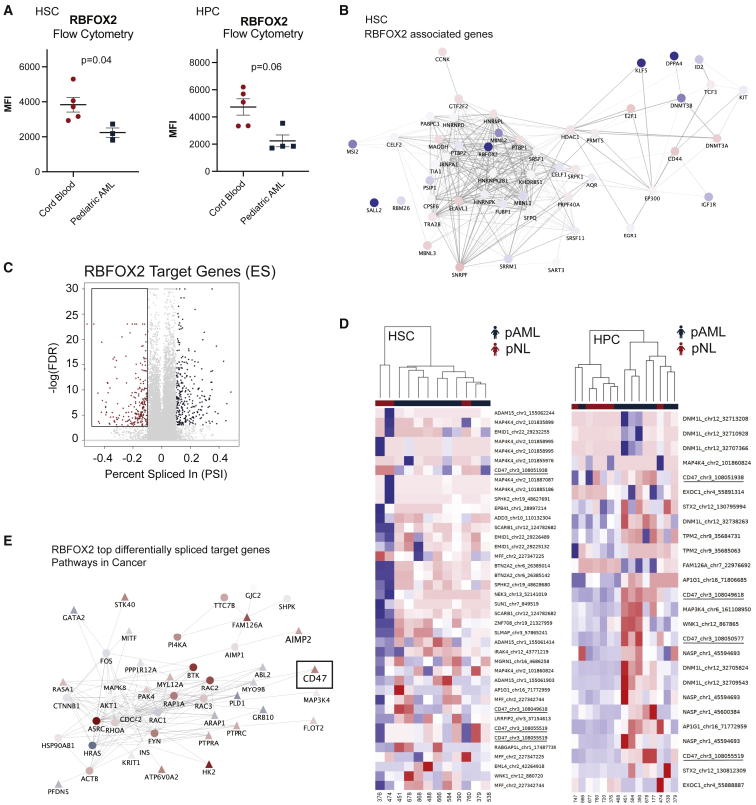
(A) RBFOX2 protein expression levels were quantified in pAML (n = 5 biological replicates) and cord-blood-derived HSCs (n = 3 biological replicates) and hematopoietic progenitor cells (HPCs; pAML, n = 5; pNL, n = 4 biological replicates) by FACS analysis as shown by MFI (Student’s t test HSC [left] p = 0.04; HPC [right] p = 0.06).
(B) RNA-seq-based cytoscape network analysis of RBFOX2 and embryonic stem cell (ESC) specific alternative splicing program-related transcripts. Nodes are colored according to log fold change.
(C) Volcano plot highlighting RBFOX2 target genes that show differential exon usage in pAML (false discovery rate, FDR < 0.05; percent spliced in, PSI > 0.1). A negative PSI reflects exon inclusion in pAML, while a positive PSI reflects ES in pAML versus pNL cells.
(D) Heatmap of exon expression levels after unsupervised clustering of RBFOX2 target genes in pAML HSCs and HPCs (n = 9 and n = 8, respectively; blue) compared with pNL (n = 3 and n = 6, respectively; red).
(E) Cytoscape network analysis of differentially spliced RBFOX2 target genes implicated in “Pathways in Cancer” in pAML HPCs. The top 200 ES events in this comparison were used as seeds for network propagation on the STRING high-confidence interactome. Differential expression log fold change is mapped to node color. Triangles depict seed list genes, and circles represent associated genes. See also Figure S5.