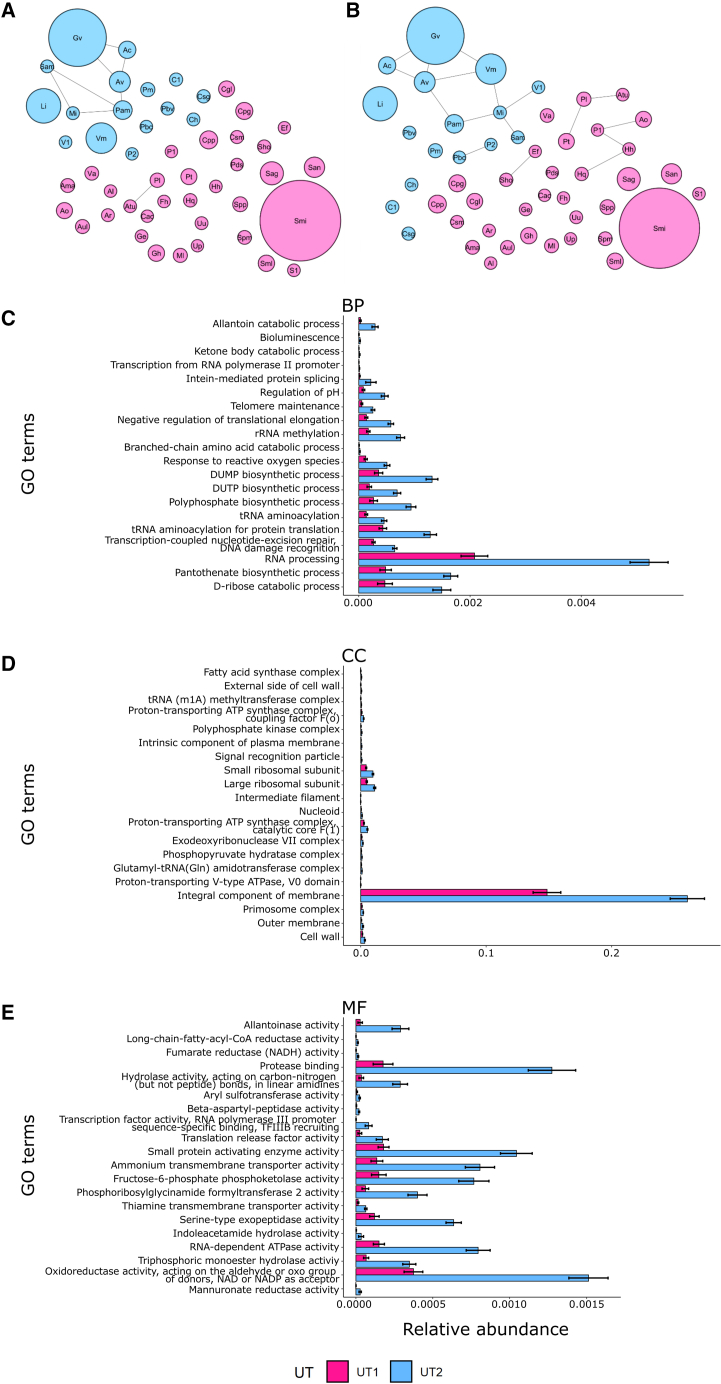
Figure 6.
SPIEC-EASI network visualizations were generated by using two inference methods to construct a microbiome association network from all the bacterial operational taxonomic units (OTUs) (117)
Each node diameter is proportional to the mean of that OTU’s relative abundance. Nodes are colored based on the urethrotype (UT1 = pink; UT2 = blue) in which the taxon is most abundant. Topology network using (A) graphical least absolute shrinkage and selection operator (Glasso), and (B) Meinshausen-Buhlmann’s neighborhood selection (MB). Edges indicate the two nodes are connected. Taxa definitions: Ac = Aerococcus christensenii; Al = Anaerococcus lactolyticus; Ama = Actinobaculum massiliense; Ao = Alloscardovia omnicolens; Ar = Actinomyces radingae; Atu = Actinomyces turicensis; Aul = Actinotignum urinale; Av = Atopobium vaginae; Cac = Cutibacterium acnes; Cgl = Corynebacterium glucuronolyticum; Ch = Corynebacterium hadale; Cpg = Corynebacterium pseudogenitalium; Cpp = Corynebacterium pyruviciproducens; Csm = Corynebacterium simulans; Csg = Corynebacterium singulare; C1 = Corynebacterium sp. NML140438; Ef = Enterococcus faecalis; Fh = Facklamia hominis; Ge = Gemella haemolysans; Gv = Gardnerella vaginalis; Hh = Haemophilus haemolyticus; Hq = Haemophilus quentini; Li = Lactobacillus iners; Mi = Mageeibacillus indolicus; Ml = Micrococcus luteus; Pam = Prevotella amnii; Pbv = Prevotella bivia; Pbc = Prevotella buccalis; Pds = Prevotella disiens; Pl = Propionimicrobium lymphophilum; Pm = Prevotella melaninogenica; P1 = Prevotella sp. oral taxon 299; P2 = Prevotella sp. S7 18; Pt = Prevotella timonensis; Sag = Streptococcus agalactiae; Sam = Sneathia amnii; San = Streptococcus anginosis group; Sho = Staphylococcus hominis; Sml = Streptococcus milleri; Smi = Streptococcus mitis; Spm = Streptococcus pneumoniae; Spp = Streptococcus pseudopneumoniae; S1 = Streptococcus sp. HMSC034E03; Up = Ureaplasma parvum; Uu = Ureaplasma urealyticum; Va = Veillonella atypica; V1 = Veillonellaceae bacterium DNF00626; Vm = Veillonella montpellierensis.
(C–E) Relative abundance of top 20 significant gene ontology (GO) terms in UT1 and UT2 generated with HUMAnN 3.0 (all p values <0.001). GO terms were mapped from the gene families in the output, and the same GO terms of different taxa were combined. Wilcoxon signed rank test was used to identify the differentially abundant GO terms between UT1 and UT2, and the GO terms were ordered by p value. GO definitions: BP = Biological process; CC = Cellular component; MF = Molecular function.