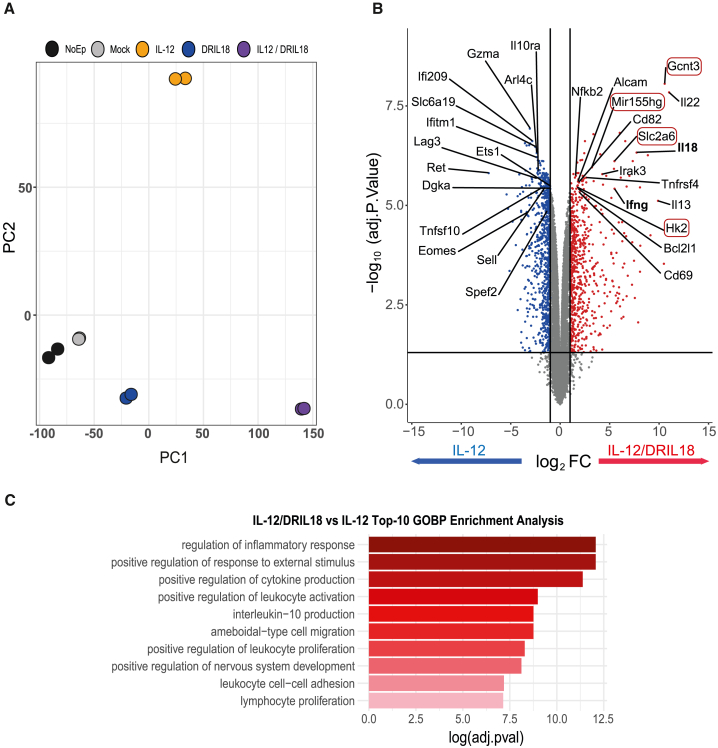
Figure 2.
RNA-seq analyses of mRNA electroporated mixed Pmel-1 cells to express scIL-12 or DRIL18
(A) Principal-component analysis of the transcriptomes of 48-h antigen preactivated Pmel-1 cells transfected with the indicated mRNAs. In the case of IL-12/DRIL18, cells were separately electroporated with either single mRNA and mixed together 1:1. RNA-transduced Pmel-1 lymphocytes were cultured for an additional 24 h before retrieving the RNA.
(B) Volcano plot analysis showing genes up-regulated or down-regulated when comparing scIL-12 mRNA with the mixture of Pmel-1 cells electroporated with scIL-12 or DRIL18. Listed genes are those considered of potential functional significance for T cells, and those whose names are surrounded by red rectangles are those that we focused on in subsequent research.
(C) Top 10 biological processes obtained in Gene Ontology (GO) enrichment analysis using the up-regulated gene list of IL-12/DRIL18 vs. scIL-12 mRNA-electroporated Pmel-1 cells (adjusted P value < 0.05 and Log2FC > 1) against “biological process” ontology. Experiments represent the data of two biological replicates. FC, fold change.
The significance is represented with asterisks (∗) according to the following values: p<0.05 (∗), p<0.01(∗∗), p<0.001(∗∗∗) and p<0.0001(∗∗∗∗).