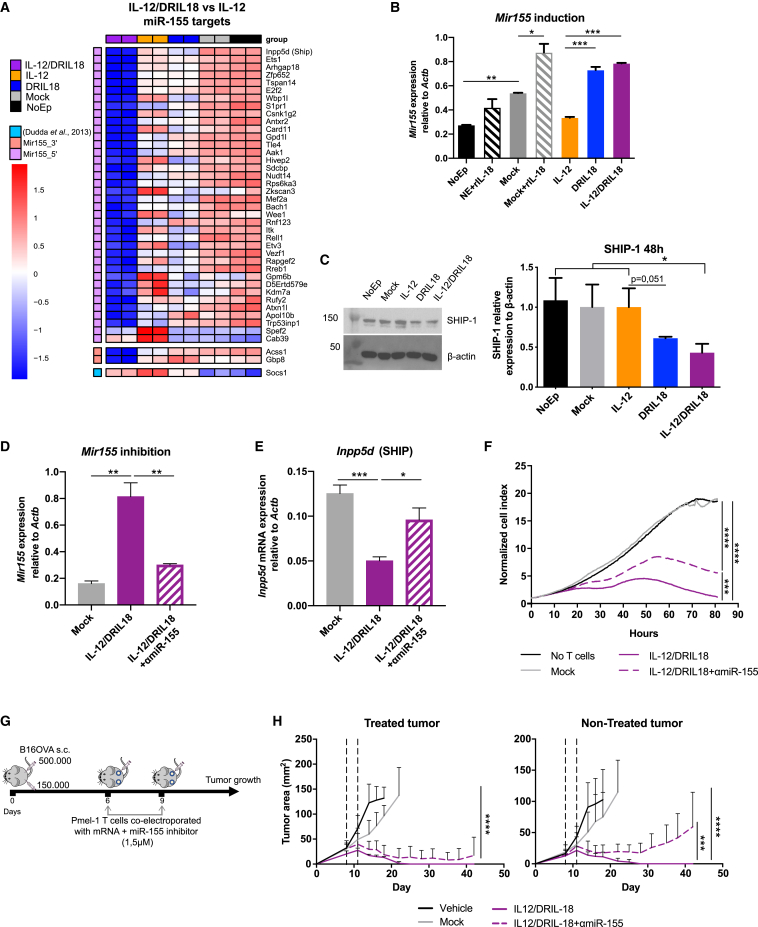
Figure 5.
Mixed scIL-12/DRIL18 electroporated Pmel-1 cells upregulate functional miR-155 that is involved in the therapeutic efficacy
(A) Heatmap showing down-regulation of the Z score log2CPM of differentially expressed genes reported as miR-155 target genes in the mixes of scIL-12/DRIL18 Pmel-1 cells. GSEA shows enrichment in miR-155-5′ targets gene set (p.val = 0.0009229154; NES = −1.654717). We added more differentially expressed (DE) genes genes that are targets of miR-155-3′ (Acss1 and Gbp8) and Socs1, a well-known target of miR-155-5′ not described in the M3 category.
(B) Confirmation of the up-regulation of Mir155 by quantitative RT-PCR (qRT-PCR).
(C) Immunoblot analysis of SHIP-1 expression in the indicated Pmel-1 mRNA-transfected cells as compared with β-actin. Relative densitometry data are shown in the bar graph.
(D) An antagomir RNA was co-electroporated to induce the degradation of miR-155 (αmiR-155) as compared with the negative control antagomir co-electroporated with IL-12/DRIL18 mRNAs.
(E) The antagomir partially rescued the mRNA expression of Inpp5d (SHIP) in the IL-12/DRIL18 pmel-1 cells as assessed by qRT-PCR.
(F) IL-12/DRIL18 pmel-1 cells co-electroporated with the antagomir degrading miR-155 exhibited less efficacious ex vivo cytotoxicity against B16-OVA as assessed by xCELLigence assays.
(G and H) Efficacy experiments in mice bearing bilateral B16-OVA tumors that show that co-electroporation of the antagomir (αmiR-155) reduced the bilateral efficacy of IL-12/DRIL18 electroporated pmel-1 cells injected to one of the tumors. Statistical comparisons were performed using one-way ANOVA (B–E) or two-way ANOVA tests (H). Experiments were repeated at least twice.
Biological duplicates were performed in experiments (A)–(F). For antitumor efficacy experiments (G and H), we randomly assigned six mice per group. Data are represented as mean ± SD. See also Figures S7 and S8.
The significance is represented with asterisks (∗) according to the following values: p<0.05 (∗), p<0.01(∗∗), p<0.001(∗∗∗) and p<0.0001(∗∗∗∗).