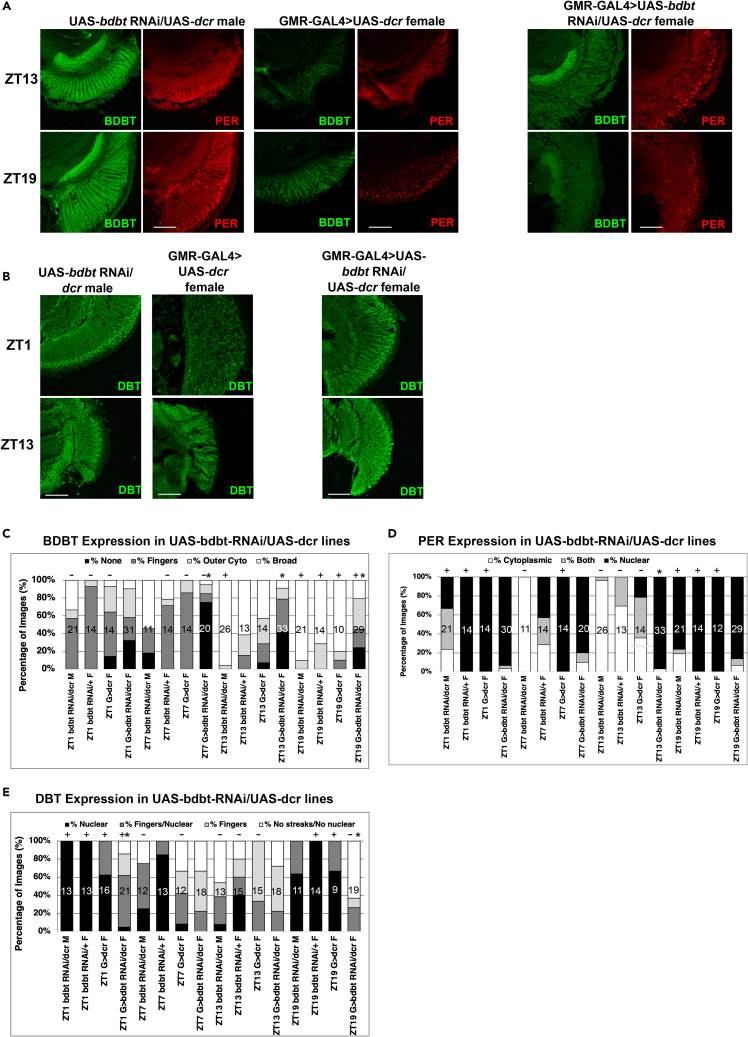
Figure 6.
Effects of BDBT RNAi on PER and DBT Localization in the Fly Eye
Fly heads were collected at ZT1, ZT7, ZT13, and ZT19, and cryosections were prepared for immunofluorescent detection of BDBT, DBT, and PER.
(A) Representative confocal Z stacks of GMR-GAL4/+>UAS-bdbt-RNAi/UAS-dcr female (knockdown), UAS-bdbt RNAi/UAS-dcr male, and GMR-GAL4>UAS-dcr female control photoreceptors at ZT13 and ZT19 for PER and BDBT localization. BDBT knockdown is supported here and also in Figure S4.
(B) Representative DBT subcellular localization in UAS-bdbt RNAi/UAS-dcr control flies, GMR-GAL4>UAS-dcr control flies, and GMR-GAL4>UAS-bdbt-RNAi/UAS-dcr flies.
(C) Graph illustrating the percentage of images scored by observers blinded to sample identity for BDBT foci expression, (D) PER localization, and (E) DBT localization. A star over a sample indicates a statistically significant different BDBT, PER, or DBT localization for GMR-GAL4>UAS-bdbt RNAi/UAS-dcr from that of any of the three control genotypes at that time point (p < 0.05 by multiple comparisons of mean ranks after Kruskal-Wallis nonparametric ANOVA analysis of each time point), while time points marked with a “+” exhibited a peak for nuclear localization (for PER or DBT) or broad expression (for BDBT) relative to those marked with a “-“ for the same genotype with statistical significance (p < 0.04 by multiple comparisons of mean ranks after Kruskal-Wallis nonparametric ANOVA analysis of each genotype). Scale bars indicate 50 μm, and the number of sections analyzed for each bar on the plot is indicated inside the bar.