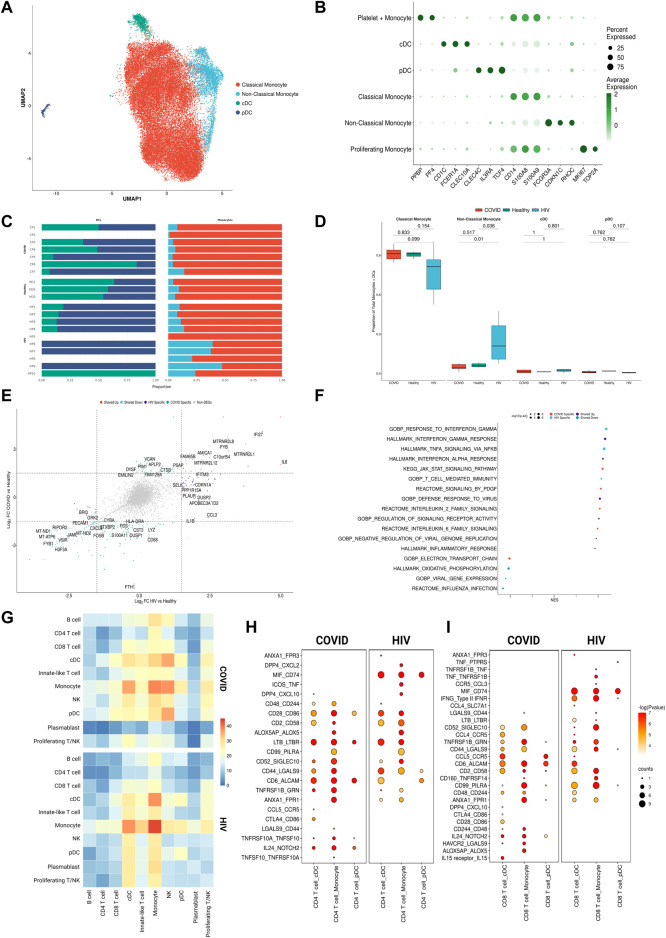
FIGURE 3.
Monocytes in COVID-19 and HIV-1 share inflammatory signatures. (A) UMAP embeddings of monocytes and DCs colored by subtype. (B) Dot plot of canonical monocyte and DC marker expression across subtypes. (C) Stacked bar plots of the relative frequency of subtypes present in each patient. (D)Box plots of the proportions of each myeloid subset across each disease condition. Proportions are computed for each patient by dividing their number of cells in each DC/monocyte subset by their total number of DCs + monocytes. p-values are computed with Wilcoxon signed-rank test with Holm-Bonferroni adjustment. (E) Double differential gene expression plot of genes that are differentially expressed between COVID-19 patients compared to healthy controls or differentially expressed between HIV-1+ patients compared to healthy controls. (F) Dot plot of enriched biological pathways from differentially expressed genes that were found to be upregulated (right, positive) or downregulated (left, negative) compared to healthy controls. (G) Heatmap of the number of receptor-ligand interactions between each cell type in COVID-19 patients (top) and HIV-1+ patients (bottom). (H) Dot plot of selected receptor-ligand interactions between CD4+ T cells and monocytes/DCs in COVID-19 patients (left) versus HIV-1+ patients (right). Color of each dot corresponds to the inverse log of the p-value of the interaction. Size of the dot corresponds to the number of patients the interaction was found to be significant in. (I) Dot plot of selected receptor-ligand interactions between CD8+ T cells and monocytes/DCs in COVID-19 patients (left) versus HIV-1+ patients (right). Color of each dot corresponds to the inverse log of the p-value of the interaction. Size of the dot corresponds to the number of patients for which the interaction was found to be significant.