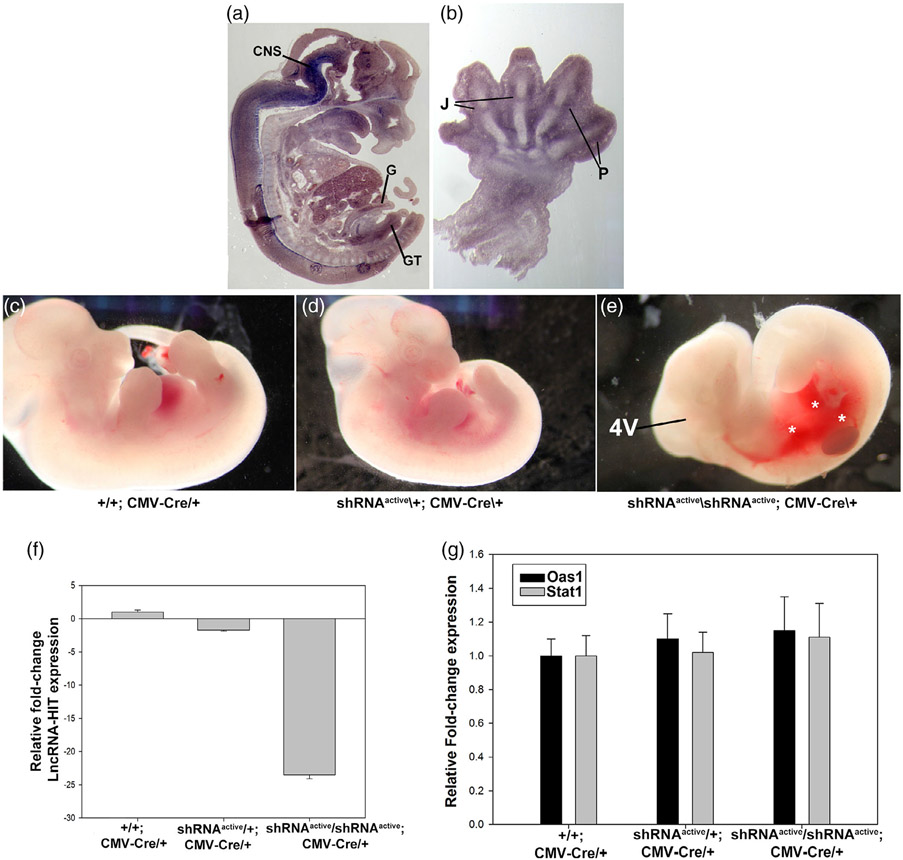
FIGURE 3.
Whole embryo activation of the LncRNA-HIT shRNA allele using CMV-Cre. (a) LncRNA-HIT is expressed in multiple tissues during development. In situ hybridization using a whole embryo sagittal section at E13.5 reveals strong expression in the developing embryo. CNS = central nervous system. G = gut epithelium. B = bladder epithelium. GT = genital tubercle. (b) Section In situ hybridization of a E13.5 forelimb limb bud reveals strong expression in the developing joint fields (J) and perichondrial (P) tissues of the digits. (c–e) E13.5 heterozygous and homozygous shRNAactive; CMV-Cre/+ embryos exhibit severe hemorrhaging (asterisks) and reductions in the fourth ventricle region of the CNS compared to either heterozygous or wild type littermate controls. (f) qRTPCR analyses of E13.5 embryos reveal >24-fold reduction in lncRNA-HIT mRNA levels between control and homozygous shRNAactive/CMV-Cre/+ embryos. (h) qRTPCR analysis of Stat1 and Oas1 expression which function as downstream indicators of a generalized interferon response reveals no differences in expression between control and shRNAactive/shRNAactive; CMV-CRE embryos. Bars in panels (f) and (g) represent the mean and standard deviation for four independent replicates