Figure 2. Transferred macrophage mitochondria are long-lived, depolarized, and accumulate reactive oxygen species, promoting cancer cell proliferation.
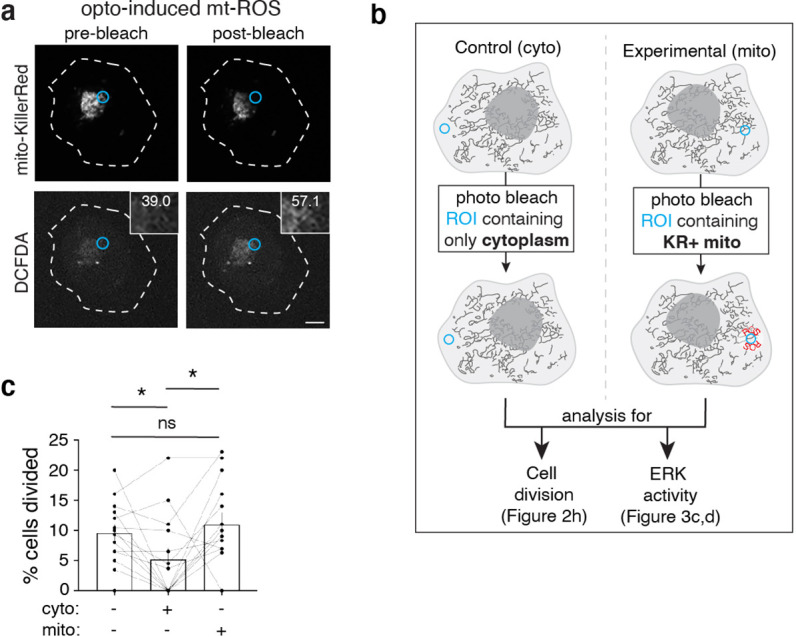
(a) Stills from time-lapse imaging depicting the longevity of the transferred mitochondria (green, arrowhead) within a 231 cell (magenta, cell outline in white). Time elapsed listed in left corner. (b) Confocal image of a mito-RFP+ 231 cell (magenta) containing macrophage mitochondria (green, arrowhead) stained with MTDR (yellow) and LysoTracker (teal). MTDR does not accumulate in 100% of donated mitochondria (N=25 cells, 5 donors). Majority (57%) of donated mitochondria do not colocalize with LysoTracker signal (N=24 cells, 4 donors). (c) Ratiometric quantification of mito-Grx1-roGFP2 biosensor mapped onto the recipient 231 cell with fire LUT (top panel). Confocal image of mito-Grx1-roGFP2-expressing 231 cell (bottom right, green and yellow) containing a macrophage mitochondria (bottom left, red, arrowhead). (d) Ratiometric measurements of the mito-Grx1-roGFP2 sensor per 231 cell (paired dots) at a region of interest containing the host mitochondrial network (host) or a transferred mitochondria (transfer). Cells were co-cultured for 24 hr (N=27 cells, 3 donors indicated in shades of gray). (e) Exogenous purified macrophage mitochondria (green) is void of mitochondrial membrane potential (MitoTracker Deep Red-negative, yellow, arrowhead) in cancer cells. (f) Cell cycle analysis of cancer cells with exogenous purified macrophage mitochondria versus sister cells that did not take up exogenous purified mitochondria, either treated with vehicle or 100 μM mitoTEMPO (mitochondrially-targeted superoxide scavenger. N=3 donors; statistics for G2/M only). (g) Schematic of optogenetic experiments to generate data in (h). Cells expressing mito-KillerRed are photobleached in a specific ROI containing either cytoplasm only (left) or mito-KillerRed+ mitochondria (right). Following photobleaching, cells are imaged over time to quantify the amount of cell division. (h) Quantification of cell division after photobleaching. Each data point is the average within a field of view (N=13 experiments), with control (cyto) and experimental (mito) data shown as paired dots per experiment. Scale bars are 10 µm. Wilcoxon matched-pairs signed rank test (d, h), two-way ANOVA (f), *p<0.05; ****p<0.0001.
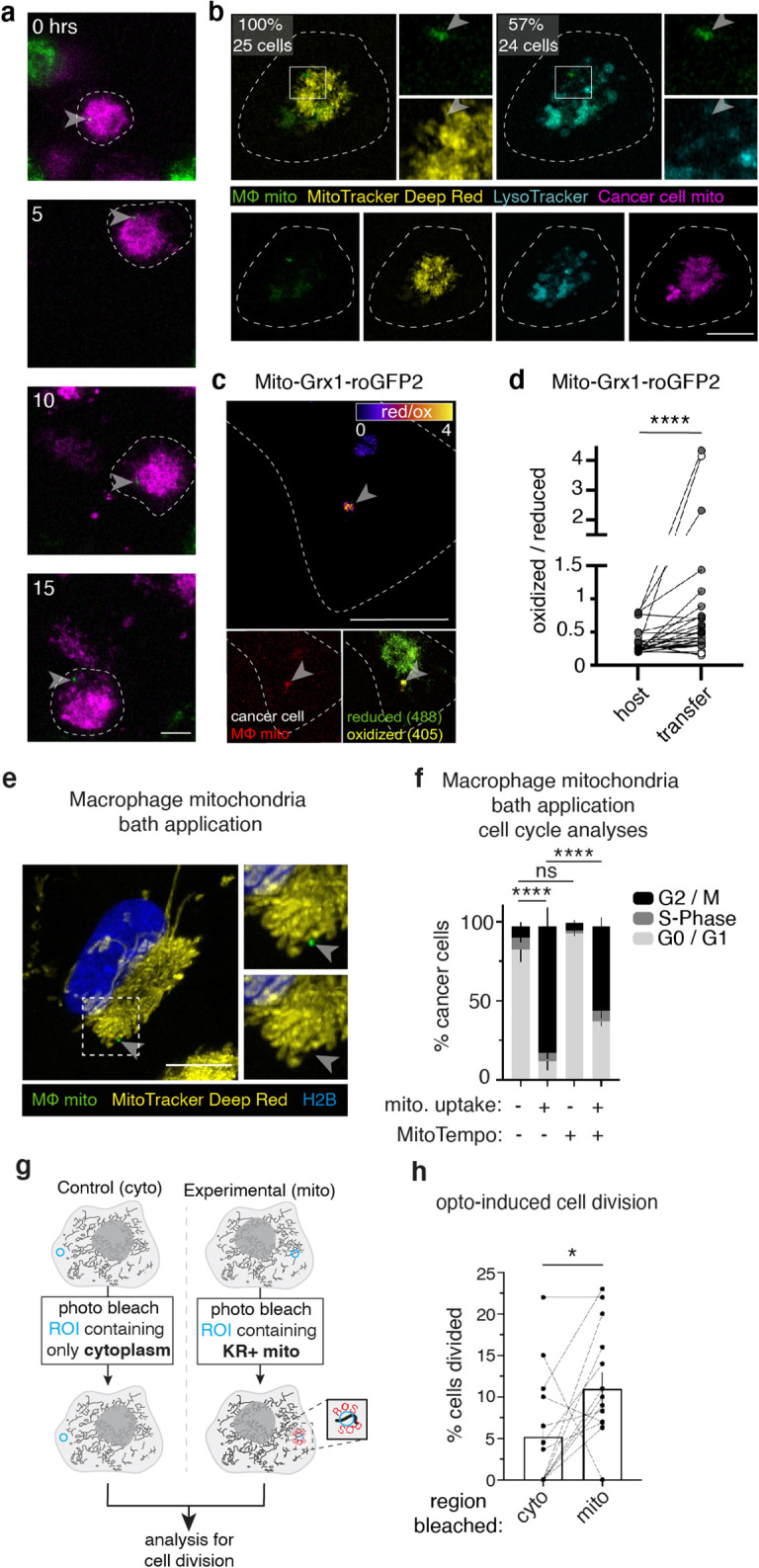
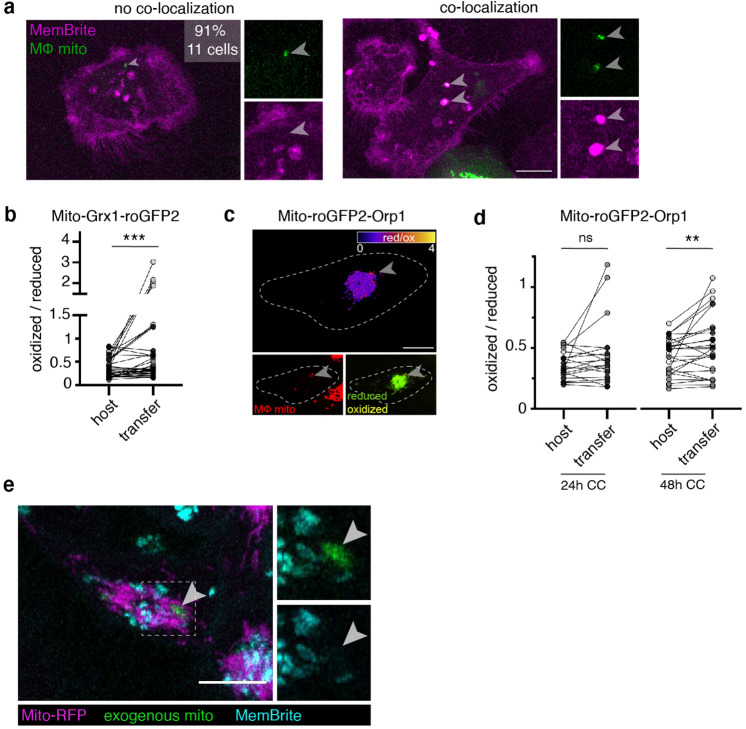
Figure 2—figure supplement 1. Transferred mitochondria accumulate reactive oxygen species, and internalized exogenous mitochondria are not encapsulated in a membrane compartment.
Figure 2—figure supplement 2. Inducing reactive oxygen species results in cancer cell proliferation.