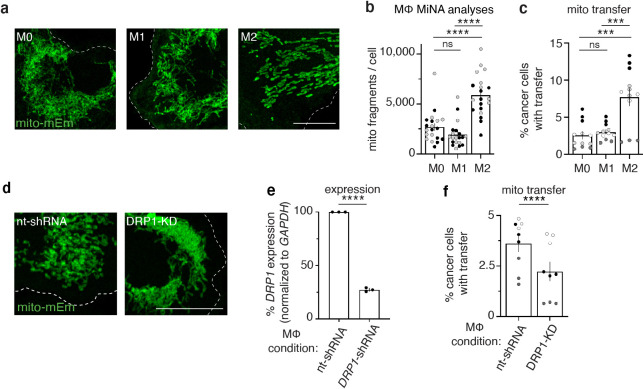
Figure 4. M2-like macrophages exhibit increased mitochondrial fragmentation and increased mitochondrial transfer to cancer cells.
(a) Representative images of mito-mEm+ macrophages that were non-stimulated (M0, left) or activated to become M1-like (middle) or M2-like (right). (b) Mitochondrial network analyses (MiNA) were used to determine number of mitochondrial fragments per cell (N=2 donors). (c) Macrophages were co-cultured with mito-RFP 231 cells for 24 hr and mitochondrial transfer was quantified with flow cytometry (N=4 donors). (d) Representative images of mito-mEm (green) macrophages in macrophages with control nt-shRNA KD and DRP1 KD. (e) q-RT-PCR of DRP1 knockdown (DRP1-KD) macrophages (N=3 donors). (f) Rates of mitochondrial transfer with control and DRP1-knockdown macrophages (N=3 donors). For all panels, individual donors are indicated as shades of gray with each cell as a data point, error bars represent SEM and scale bars are 10 µm. Two-way ANOVA (b, c), unpaired t-test (e, f), ***p<0.001; ****p<0.0001.
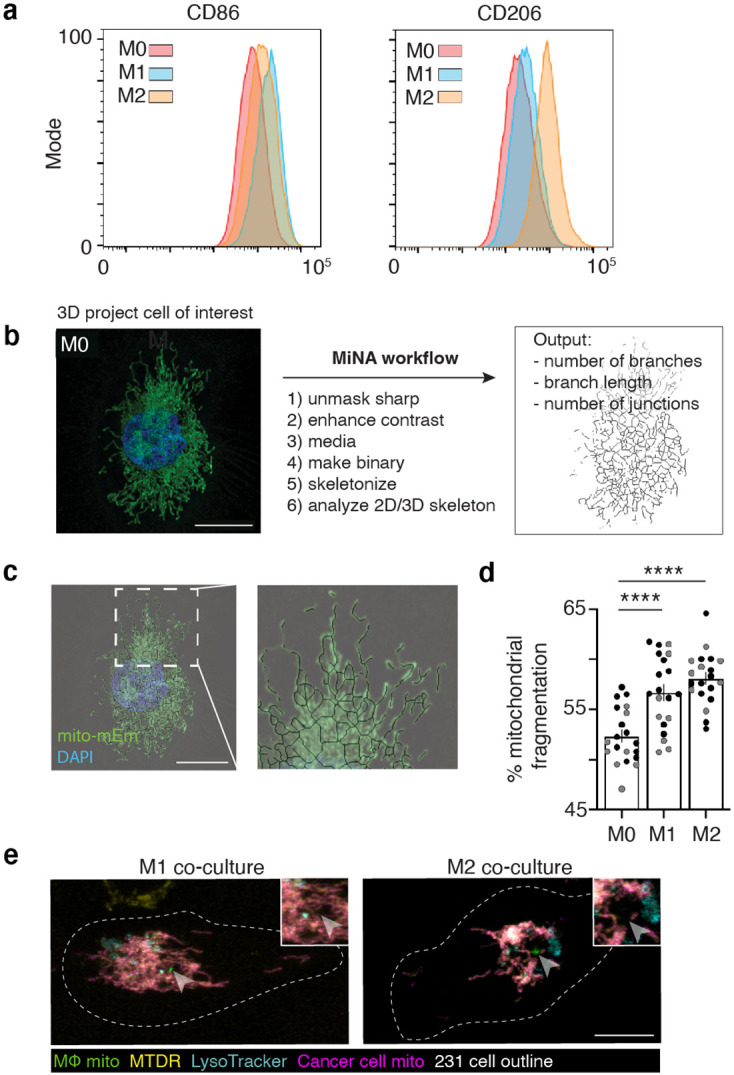
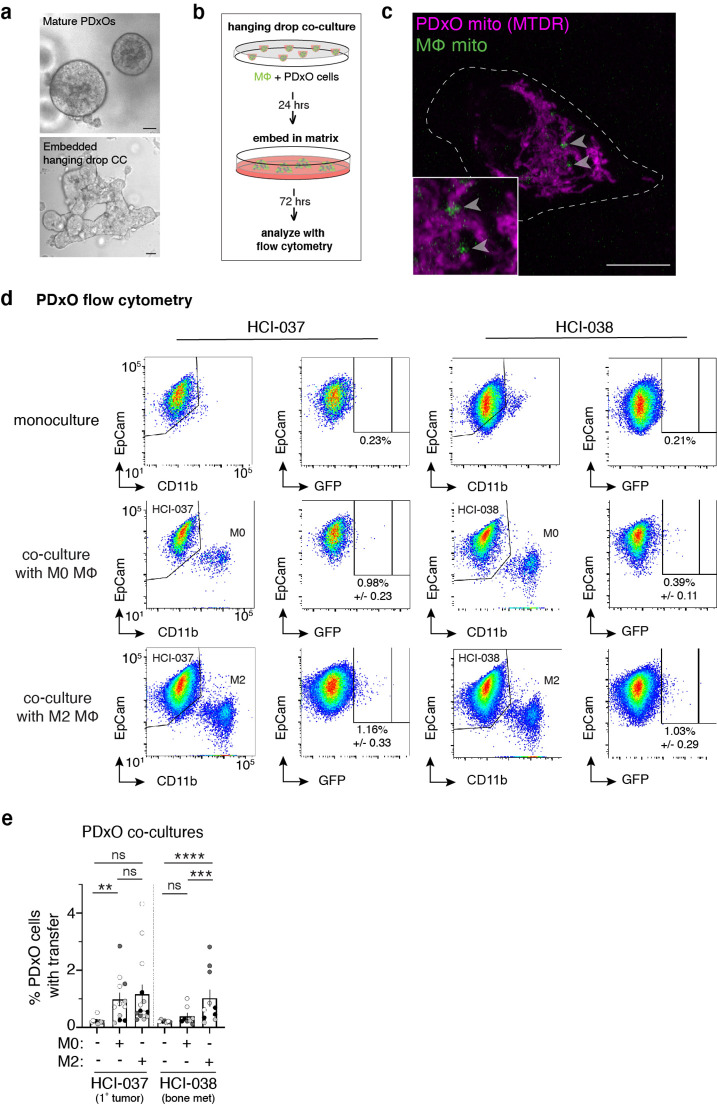
Figure 4—figure supplement 1. M2-like macrophages exhibit increased mitochondrial transfer to cancer cells.