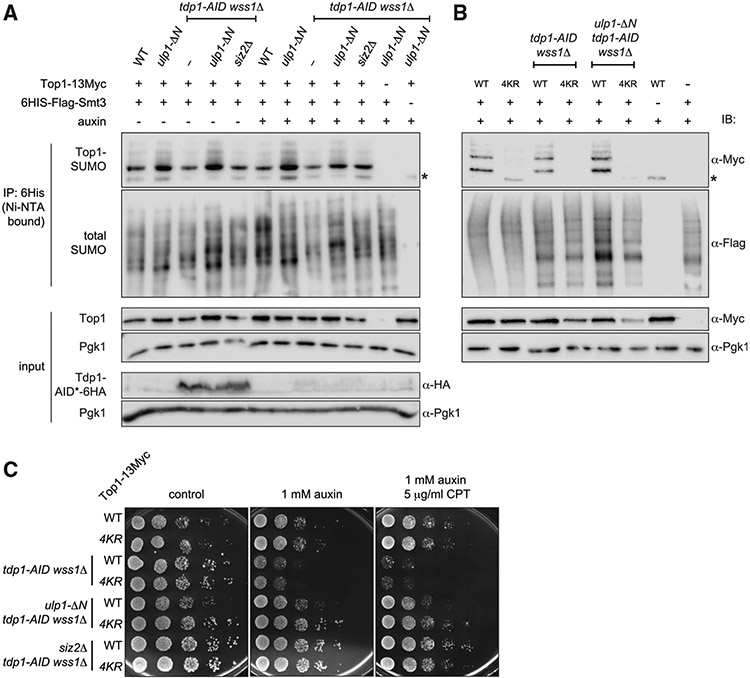
Figure 3. Top1 sumoylation in ulp1-ΔN and siz2Δ.
(A) Relative levels of sumoylated Top1 evaluated by the SUMO assay. SMT3 (gene coding for yeast SUMO) and TOP1 were tagged at genomic loci. Asterisks indicate unmodified Top1-13Myc non-specifically bound to Ni-NTA beads. Tdp1-AID*- HA was depleted by 6 h auxin treatment (note: Tdp1 is 6HA-tagged in tdp1-AID background only).
(B) Sumoylation status of Top1-13Myc or Top1-4KR-13Myc mutants. The SUMO assay was performed as in (A). TOP1-13Myc or top1-4KR-13Myc (K65R, K91R, K92R, K600R) replace the genomic copy of TOP1.
(C) CPT sensitivity of non-sumoylated Top1 mutants. Spot assays were plated on YEPD media and imaged 48 h post-plating.