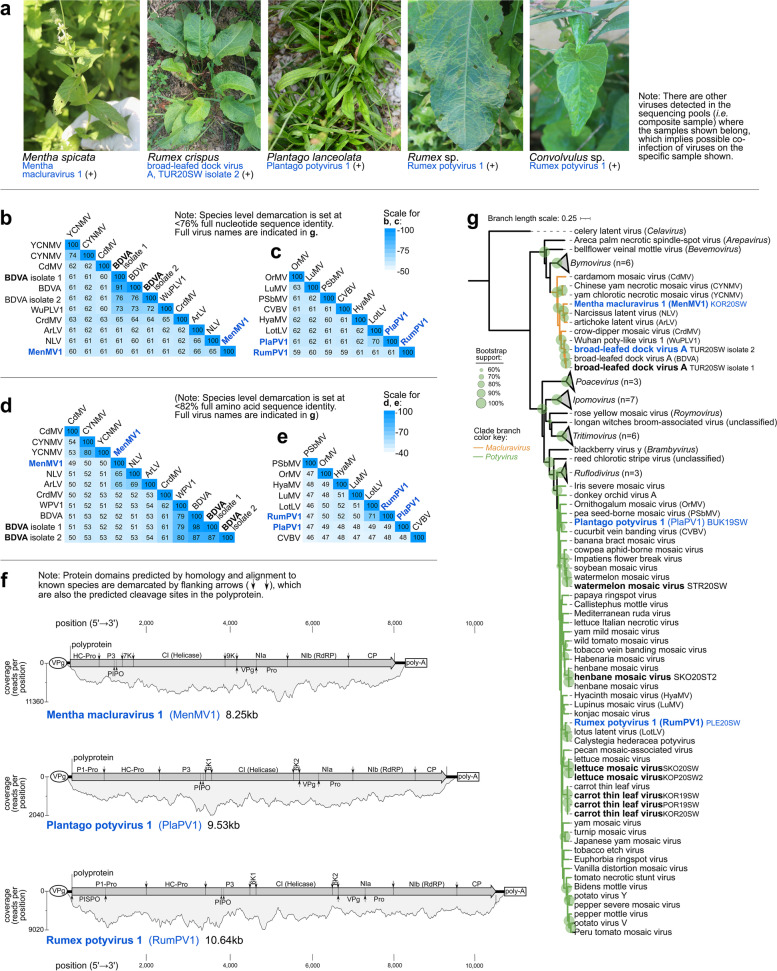
Fig. 8.
Characteristics of (+)ssRNA viruses from family Potyviridae from the present study. a Selected photographs of samples that tested positive in RT-PCR assays for the viruses presented herewith. b–e Heatmaps of the pairwise identities of Macluravirus and Potyvirus species based on alignment and comparison of full genome nucleotide sequences (b, c) and based on full polyprotein amino acid sequence (d, e). f Genome organization with predicted cleavage sites and protein domains, as well as read coverage for novel potyviruses from this study. g Maximum likelihood phylogenetic tree based on the alignment of conserved RdRp amino acid sequence of RefSeq species from the Potyviridae family. Branch length scale represents amino acid substitution per site. Virus names and acronyms in blue bold font are the novel viruses, while those in black bold font are known viruses from the present study. Indicated after the acronym are the isolate IDs of the viruses. Full virus names of those abbreviated in the pairwise identity matrices (b–e) can be found in the genome organization and phylogenetic trees (f–g)