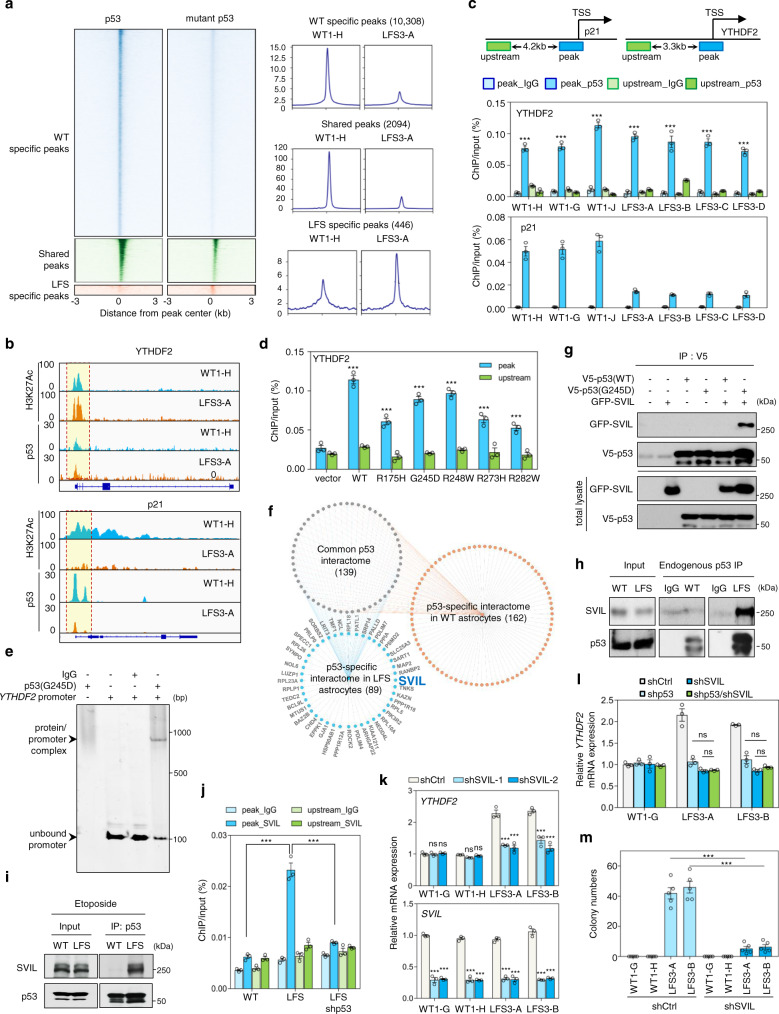
Fig. 3. Genome occupancy and interactome studies reveal that mutant p53 cooperates with SVIL in regulating YTHDF2 expression.
a Heatmaps (left panels) depict the p53/mutant p53 genomic occupancies of p53-specific, mutant p53-specific, and shared peaks within 3 kb of peak centers according to p53 ChIP-seq. Composite plots (right panels) show normalized p53 and mutant p53 density distribution at promoters of genes within p53-specific, mutant p53-specific, and shared peaks. b Integrative Genomics Viewer (IGV) track views of H3K27ac, p53, and mutant p53 genome occupancy over p21 and YTHDF2 promoter regions in multiple WT and LFS astrocytes. c ChIP-qPCR validation of p53 and p53(G245D) binding peaks at the identified YTHDF2 promoter. p53/mutant p53 binding peak (blue) and upstream non-p53/mutant p53 binding regions (green) used for ChIP-qPCR validation (n = 3 biologically independent samples). d ChIP-qPCR indicates that p53 and various p53 mutants bind to the YTHDF2 promoter but not the upstream non-p53/mutant p53 binding region (n = 3 biologically independent samples). e EMSA demonstrates direct p53(G245D) binding to the YTHDF2 promoter. f Snapshot of p53 and mutant p53 interaction network. Connectivity map between p53 and mutant p53 interactomes in WT and LFS astrocytes. g p53(G245D) but not p53 interacts with SVIL exogenously. h Endogenous interaction between p53(G245D) and SVIL in LFS astrocytes. i Etoposide-induced WT p53 activation does not alter the interaction of p53(G245D) and SVIL in LFS astrocytes. j ChIP-qPCR indicates that knockdown of p53(G245D) hampers SVIL binding on the YTHDF2 promoter in LFS astrocytes (n = 3 biologically independent samples). k RT-qPCR analysis shows decreased YTHDF2 mRNA expression upon SVIL knockdown in LFS astrocytes but not WT astrocytes (n = 3 biologically independent samples). l RT-qPCR analysis demonstrates comparable YTHDF2 mRNA expression upon depletion of mutant p53, SVIL, and mutant p53/SVIL in LFS astrocytes (n = 3 biologically independent samples). m In vitro AIG assay demonstrates decreased colony numbers upon SVIL depletion in LFS astrocytes (n = 5 biologically independent samples). The results are representative of at least three independent experiments (e, g–i). The data are presented as the mean ± SEM; two-way ANOVA with Bonferroni’s multiple comparison test (c, d, j–m). ***P < 0.001. ns not significant. Source data and exact P values are provided in the Source Data file.