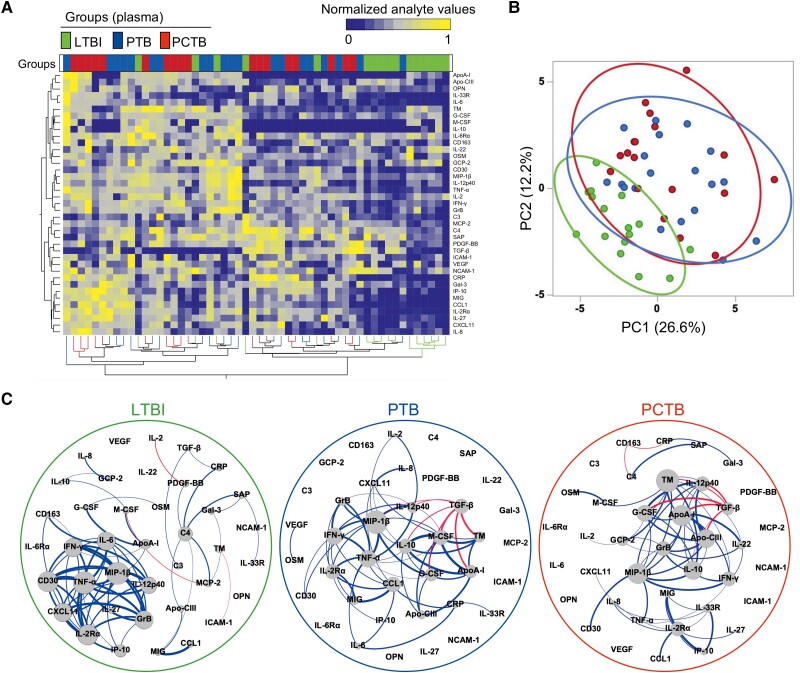
Figure 1.
Analyte profiles in the different tuberculosis (TB) groups at baseline. A, Non-supervised 2-way hierarchical cluster analysis (Ward method) was employed to evaluate the TB groups using the 39 measured analytes (see Materials and Methods for expansions of abbreviations of analytes). TB status (pericardial TB [PCTB] in red, pulmonary TB [PTB] in blue, and latent TB infection [LTBI] in green) of each patient is indicated at the top of the dendrogram. Data are depicted as a heatmap colored from minimum to maximum normalized values for each marker. B, Principal component analysis on correlations based on the 39 analytes was used to explain the variance of the data distribution in the cohort. Each dot represents a participant. The 2 axes represent principal components 1 (PC1) and 2 (PC2). Their contribution to the total data variance is shown as a percentage. C, Analyte network analysis (Fruchterman-Reingold algorithm) in plasma of LTBI, PTB, and PCTB participants. Size of nodes indicates the number of connections. Size of edges indicates the Spearman r value (only r >0.6 was included). Blue lines: positive correlation. Red lines: negative correlation.