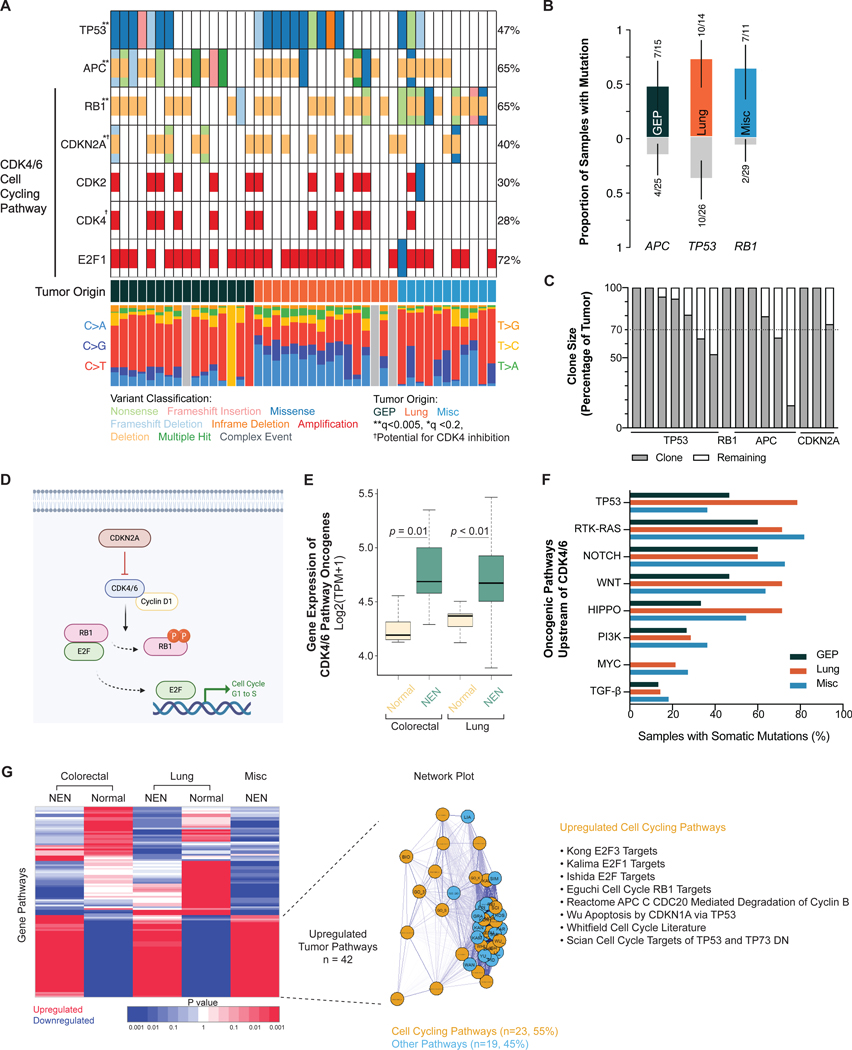
Figure 3. Significantly mutated genes and CDK4/6 cell cycling pathway dysregulation.
(a) Oncoplot showing significantly mutated genes marked by asterisk(s) per dNdScv method. Low-pass WGS n = 43; WES n = 40.
(b) TP53 mutations were enriched in lung, APC in gastroenteropancreatic, and RB1 in non-lung/non-GEP neoplasms (p < 0.05, pairwise and groupwise Fisher exact test). N = 40.
(c) EXPANDS analysis showing the clone size of each significantly mutated gene. Dotted line shows the 70% threshold often used to define a founder clone.
(d) Schematic of CDK4/6 cell cycling pathway.
(e) Boxplot showing gene expression levels of the CDK4/6 pathway oncogenes (CCND1, CCDN2, CCND3, CCNE1, CDK2, CDK4, CDK6, E2F1, E2F3) between neoplasms and normal tissue. Mean ± SD (colorectal normal: 4.26 ± 0.15, n = 7; colorectal tumor: 4.77 ± 0.35, n = 8; lung normal: 4.34 ± 0.11, n = 9; lung tumor: 4.71 ± 0.48, n = 17). Statistical test: Holm-Bonferroni method.
(f) Frequency of mutations in canonical oncogenic pathways upstream of CDK4/6 pathway.
(g) Unsupervised clustering heatmap of gene pathways. Gene pathways universally upregulated by all neoplasms compared to normal tissue (n = 42) comprised mostly of cell cycling pathways (n = 23).
Abbreviations: WGS, whole genome sequencing. WES, whole exome sequencing. GEP, gastroenteropancreatic. NEN, neuroendocrine neoplasm. Misc, miscellaneous.