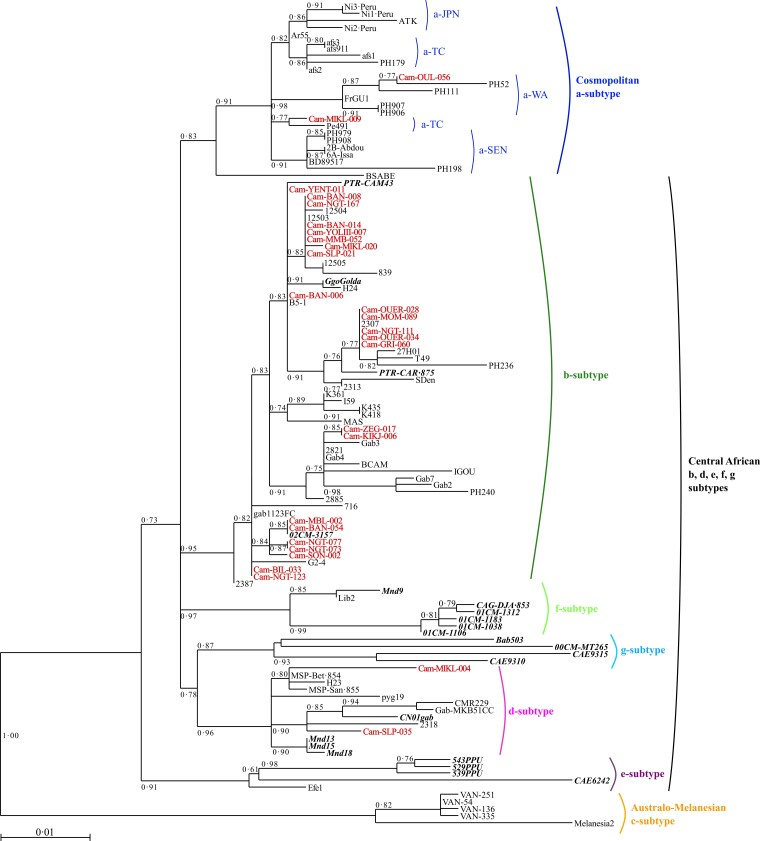
Figure 1.
Phylogenetic tree generated by the maximum likelihood method with a 522-bp fragment of the env gene. Phylogenetic comparisons were performed with the 522-nucleotide env gp21 gene fragment obtained from 113 human T-cell leukemia virus 1 (HTLV-1) isolates, including the 27 sequences from this study (in red) and 86 previously published sequences. The GenBank accession numbers of the new sequences from the rural population of Cameroon are ON130946–ON130972. Simian T-cell leukemia virus 1 (STLV-1) strains are shown in bold italic typeface. The phylogeny was derived by the maximum likelihood method with the generalized time reversible (GTR) model. Horizontal branch lengths are drawn to scale, with the bar indicating 0.01 nucleotide replacements per site. Maximum posterior probabilities were calculated and reported on the maximum likelihood tree (threshold value ≥0.50). a-TC, a-JPN, a-SEN, a-WA, correspond to the transcontinental, Japanese, Senegalese, and West African clades of the a-genotype, respectively.