Figure 1.
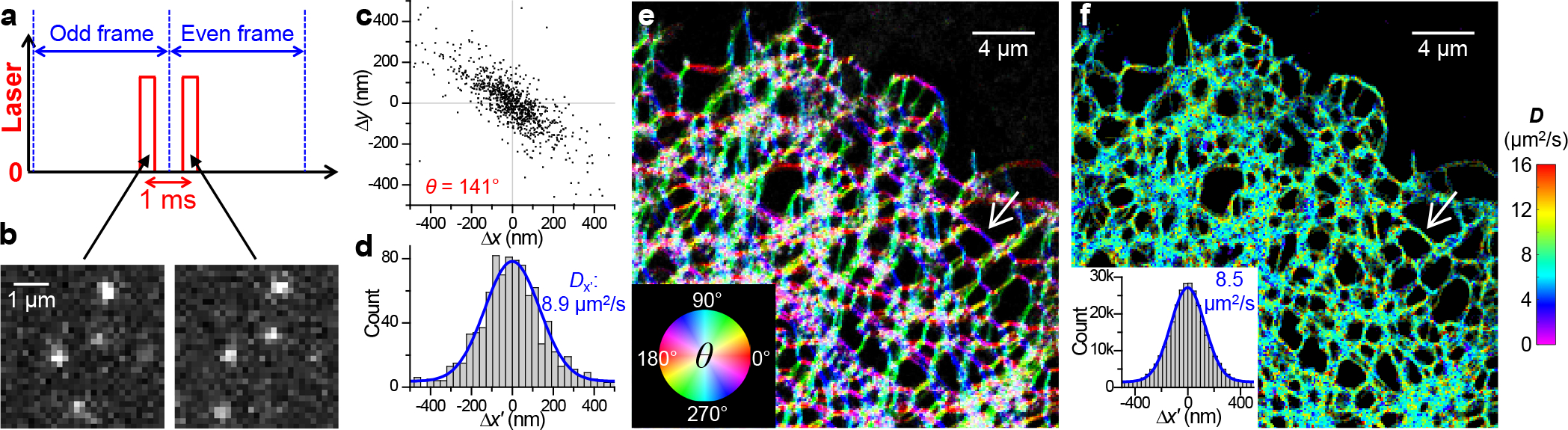
High-content single-molecule statistics and super-resolution mapping of intraorganellar diffusivity. (a) Schematics: Tandem excitation pulses of τ = 500 μs duration are applied across paired camera frames at a Δt = 1 ms center-to-center separation, and this scheme is repeated ~104 times to enable local statistics. (b) Example single-molecule images captured in a tandem frame pair, for ER-Dendra2 diffusing in the ER lumen of a live COS-7 cell. (c) Two-dimensional plot of the vectorial 1-ms single-molecule displacements accumulated for an ER-tubule segment [white arrows in (e,f)], from which a principal direction θ of 141° is assessed. (d) Distribution of the displacements in (c) projected along θ. Blue curve: maximum likelihood estimation (MLE) fit to a one-dimensional diffusion model, yielding D = 8.9 μm2/s. (e) Color map presenting the calculated local θ across the imaged COS-7 cell. (f) pSMdM super-resolution D map via local fitting along the local principal directions. Inset: Pooled distributions of the 3.1×105 single-molecule displacements in the sample, each projected along its local θ. Blue curve: MLE fit yielding D = 8.5 μm2/s.
