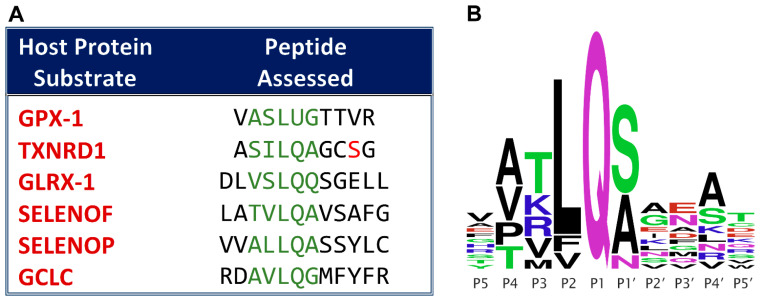
Figure 1.
(A) Synthetic peptides used for Mpro cleavage assessment of previously predicted sites from the host proteins listed [27]. The letters shown in green correspond to the P4 through P1′ positions that are most important for recognition by the enzyme. The red S in TXNRD1 is mutated from a selenocysteine in the wild type protein. (B) Consensus sequence logo for known SARS-CoV-2 Mpro cleavage sites in viral nonstructural proteins. Cleavage occurs between the conserved glutamine (Q) in the P1 position and the adjacent residue in the P1′ position. The letters stacked vertically in a given position (representing amino acid residues in single letter protein code) show the natural variation tolerated by the protease active site in that position. Simplified from Figure 1 in [27].