Figure 2.
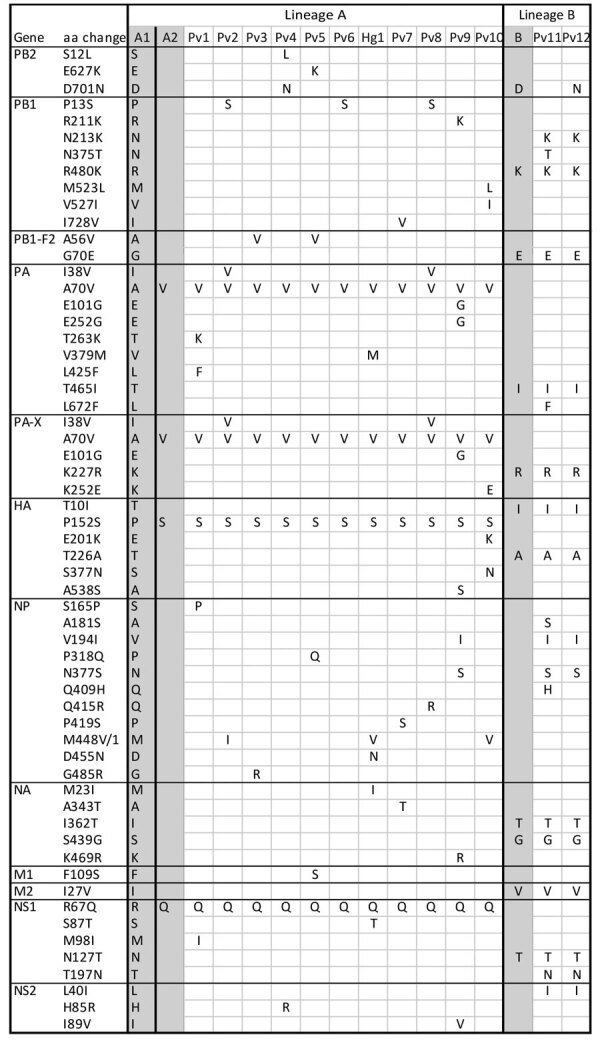
Amino acid changes in highly pathogenic avian influenza A(H5N1) viruses from New England birds and seals, United States. Each single-nucleotide polymorphism (SNP) that resulted in an amino acid change within ≥1 seal-derived sequence is shown. SNPs were observed in 12 H5N1 virus genes resulting in amino acid changes in corresponding proteins: PB2, PB1, PB1-F2, PA, PA-X, HA, NP, NA, M1, M2, NS1, and NS2. All avian virus reference sequences are shaded gray. A/Sanderling/MA/CW_22–112 (H5N1) (GISAID database, https://www.gisaid.org) (labeled A1) was used as a reference for first wave lineage A sequences; A/common eider/MA/TW_22–1400 (H5N1) (labeled A2) was used as a reference for second wave lineage A sequences; and A/common tern/MA/20220612_1 (H5N1) (labeled B) was used as a reference for lineage B sequences. Four aa differ between first and second wave lineage A viruses and 10 aa differ between first wave lineage A and lineage B. Second wave lineage A and lineage B seal-derived virus sequences, sampling date, and sampling location in Maine, USA, are indicated for each seal as follows: Pv1, MME22-112, 2022 Jun 22, Wells; Pv2, MME22-117, 2022 Jun 24, Yarmouth; Pv3, MME22-121, 2022 Jun 26, Georgetown; Pv4, MME22-122, 2022 Jun 27, New Harbor; Pv5, MME22-131, 2022 Jun 28, Harpswell; Pv6, MME22-133, 2022 Jun 29, S. Portland; Hg1, MME22-144, 2022 Jul 1, Phippsburg; Pv7, MME22-150, 2022 Jul 2, Westport; Pv8, MME22-155, 2022 Jul 2, Falmouth; Pv9, MME22-198, 2022 Jul 9, Wells; Pv10, MME22-230, 2022 Jul 15, Kennebunkport; Pv11, MME22-191, 2022 Jul 8, Harpswell; Pv12, MME22-195, 2022 Jul 9, Harpswell. HA, hemagglutinin; Hg, gray seal; M, matrix; NA, neuraminidase; NP, nucleoprotein; NS, nonstructural; PA, polymerase acidic; PB, polymerase basic; Pv, harbor seal.
