Extended Data Fig. 8 |. Comparison of microglial pseudotime DEGs.
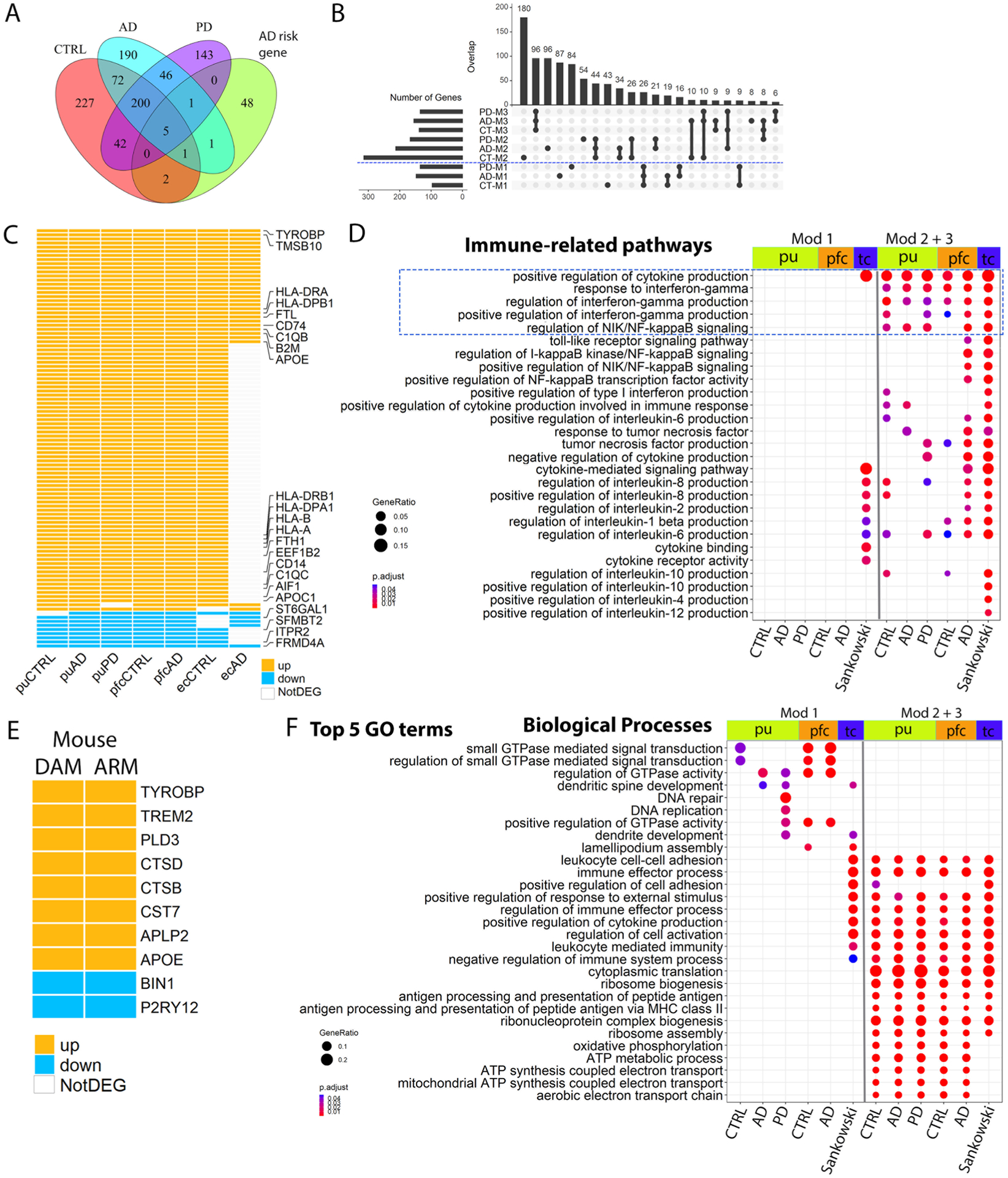
a, Venn diagram showing the overlap between pseudotime DEGs of control, AD and PD microglia with AD-risk genes (hypergeometric test). Pseudotime DEGs are genes whose expression significantly associated with pseudotime progression (generalized addictive model, FDR-adjusted P value < 0.05). b, UpSet plot showing the overlap between control, AD and PD microglial pseudotime gene coexpression modules from putamen microgla. c, Heatmap showing pseudotime DEGs shared by human activated microglia from the putamen (pu) of cognitively normal controls, AD and PD samples, from prefrontal cortex (pfc) of the control and AD samples and from the entorhinal cortex (ec) of the control and AD samples. d, GO terms related to immune functions enriched in the microglia pseudotime DEGs. e, Heatmap showing pseudotime DEGs shared by the mouse activated microglia DAM and ARM. f, Top 5 GO terms in the biological process category enriched in the microglia pseudotime DEGs. Pathways with FDR-adjusted P value < 0.05 (hypergeometric test) and at least five query genes were considered statistically significant. DAM: Disease-associated microglia; ARM: activated response microglia. UP: upregulated during pseudotime progress (module 2 and 3 genes). Down: downregulated during pseudotime progress (module 1 genes). Mod 1: module 1 genes; Mod 2 + 3: module 2 and 3 genes combined.
