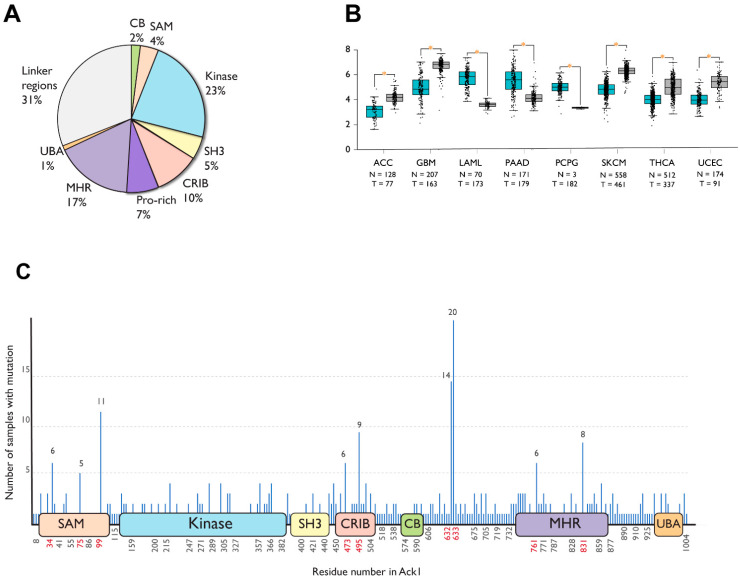
Figure 5.
Cancer-associated mutations in Ack1 in the COSMIC database. (A) Distribution of nonsynonymous mutations. SAM, sterile alpha motif; CRIB, Cdc42, and Rac-interactive binding (CRIB) domain; CB, clathrin-binding motif; proline-rich region; MHR, Mig6 homology region; UBA, ubiquitin-associated domain (B) Ack1 expression in normal (blue) vs. tumor (gray) tissues. ACC: adrenocortical carcinoma, GBM: glioblastoma multiforme, LAML: acute myeloid leukemia, PAAD: pancreatic adenocarcinoma, PCPG: pheochromocytoma, SKCM: skin cutaneous melanoma, THCA: thyroid carcinoma, UCEC: uterine corpus endometrial carcinoma. (C) Locations of cancer-associated mutations outlined in the linear structure of Ack1. SAM, sterile alpha motif; NES, nuclear export signal; CRIB, Cdc42, and Rac-interactive binding (CRIB) domain; clathrin-binding motif; proline-rich region; MHR, Mig6 homology region; UBA, ubiquitin-associated domain. Each horizontal line represents a mutation, with the number of mutations positively correlated with the length depicted on the y-axis. The x-axis shows the location of the mutations. Highly mutated residues are shown in red. Note that the numbering convention used in COSMIC is based on an isoform different from the canonical one (UniProtKB: A0A5F9ZHL4), and that the figure uses the isoform 1 (UniProtKB: Q07912) convention.