Abstract
Background
Antigen lateral flow devices (LFDs) have been widely used to control SARS-CoV-2. We aimed to improve understanding of LFD performance with changes in variant infections, vaccination, viral load, and LFD use, and in the detection of infectious individuals.
Methods
In this diagnostic study, paired LFD and RT-PCR test results were prospectively collected from asymptomatic and symptomatic participants in the UK between Nov 4, 2020, and March 21, 2022, to support the National Health Service (NHS) England's Test and Trace programme. The LFDs evaluated were the Innova SARS-CoV-2 Antigen Rapid Qualitative Test, the Orient Gene Rapid Covid-19 (Antigen) Self-Test, and the Acon Flowflex SARS-CoV-2 Antigen Rapid Test (Self-Testing). Test results were collected across various community testing settings, including predeployment testing sites, routine testing centres, homes, schools, universities, workplaces, targeted community testing, and from health-care workers. We used multivariable logistic regression to analyse LFD sensitivity and specificity using RT-PCR as a reference standard, adjusting for viral load, LFD manufacturer, test setting, age, sex, test assistance, symptom status, vaccination status, and SARS-CoV-2 variant. National contact tracing data from NHS Test and Trace (Jan 1, 2021, to Jan 11, 2022) were used to estimate the proportion of transmitting index patients (with ≥1 RT-PCR-positive or LFD-positive contact) potentially detectable by LFDs (specifically Innova, as the most widely used LFD) with time, accounting for index viral load, variant, and symptom status.
Findings
We assessed 75 382 pairs of LFD and RT-PCR tests. Of these, 4131 (5·5%) were RT-PCR-positive. LFD sensitivity versus RT-PCR was 63·2% (95% CI 61·7–64·6) and specificity was 99·71% (95% CI 99·66–99·74). Increased viral load was independently associated with being LFD positive (adjusted odds ratio [aOR] 2·85 [95% CI 2·66–3·06] per 1 log10 copies per mL increase; p<0·0001). There was no evidence that LFD sensitivity differed for delta (B.1.617.2) infections versus alpha (B.1.1.7) or pre-alpha (B.1.177) infections (aOR 1·00 [0·69–1·45]; p=0·99), whereas omicron (BA.1 or BA.2) infections appeared more likely to be LFD positive (aOR 1·63 [1·02–2·59]; p=0·042). Sensitivity was higher in symptomatic participants (68·7% [95% CI 66·9–70·4]) than in asymptomatic participants (52·8% [50·1–55·4]). Among 347 374 unique index patients with probable onward transmission, 78·3% (95% CI 75·3–81·2) were estimated to have been detectable with LFDs (Innova), and this proportion was mostly stable with time and for successive variants. Overall, the estimated proportion of infectious index patients detectable by the Innova LFD was lower in asymptomatic patients (57·6% [53·6–61·9]) versus symptomatic patients (79·7% [76·7–82·5]).
Interpretation
LFDs remained able to detect most SARS-CoV-2 infections throughout vaccine roll-out and across different viral variants. LFDs can potentially detect most infections that transmit to others and reduce the risk of transmission. However, performance is lower in asymptomatic individuals than in symptomatic individuals.
Funding
UK Health Security Agency, the UK Government Department of Health and Social Care, National Institute for Health Research (NIHR) Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance, and the University of Oxford NIHR Biomedical Research Centre.
Introduction
Early detection of symptomatic and asymptomatic SARS-CoV-2 infections has been a key control measure during the COVID-19 pandemic. Rapid point-of-care antigen detection lateral flow devices (LFDs) have been widely used in testing in the UK,1, 2 including, at different times, for population-wide asymptomatic screening and for screening in specific groups, such as health-care workers, school-age children, and populations with increased COVID-19 incidence. LFDs have also been used to allow contacts of infected patients to continue to work or attend education, as well as prior to travel, attending events, and visiting residential care facilities.
Research in context.
Evidence before this study
Antigen lateral flow devices (LFDs; ie, rapid antigen detection devices) have been widely used for SARS-CoV-2 testing. However, due to their imperfect sensitivity when compared with PCR and absence of a widely available gold standard proxy for infectiousness, the performance and use of LFDs has been a source of debate. We conducted a literature review in PubMed, bioRxiv, and medRxiv for all studies examining the performance of LFDs published between Jan 1, 2020, and Oct 31, 2022. We used the search terms “SARS-CoV-2”/“COVID-19” and “antigen”/“lateral flow test”/“lateral flow device”. Multiple studies have examined the sensitivity and specificity of LFDs, including several systematic reviews. However most studies are based on pre-alpha infections. Large studies examining test accuracy for different variants, including delta and omicron, and after vaccination are scarce.
Added value of this study
In this study of the national LFD evaluation programme in the UK, we compared the performance of three different LFDs relative to RT-PCR in various settings. Compared with RT-PCR testing, sensitivity was 63·2% (95% CI 61·7–64·6) overall, and 71·6% (69·8–73·4) in unselected community-based testing. Specificity was 99·71% (95% CI 99·66–99·74). In a multivariable model, LFDs were more likely to be positive as viral load increased. Additionally, LFD sensitivity was similar during the periods of estimated alpha (B.1.1.7) and pre-alpha (B.1.177) infections and delta (B.1.617.2) infections, but increased during the omicron (BA.1 or BA.2) period (relative to the alpha and pre-alpha period). We found no independent association between sensitivity and vaccination status. Sensitivity was higher in symptomatic participants (68·7% [66·9–70·4]) than in asymptomatic participants (52·8% [50·1–55·4]). Using national contact tracing data, we estimated that 78·3% (75·3–81·2) of index patients with probable onward transmission (ie, with one or more RT-PCR-positive or LFD-positive contact) were detectable with the Innova LFD on the basis of performance in unselected community-based testing. Symptomatic index patients were more likely to be detected than asymptomatic index patients due to higher viral loads and better LFD performance at a given viral load. The estimated proportion of detectable index patients remained mostly stable with time and for alpha, delta, and omicron infections, with a slight increase in the estimated proportion of asymptomatic index patients detectable during omicron.
Implications of all the available evidence
Our data show that LFDs detect most SARS-CoV-2 infections, with findings broadly similar to those summarised in previous meta-analyses. We show that LFD performance has been generally consistent throughout different variant-dominant phases of the pandemic and after the roll-out of vaccination. LFDs can detect most infections that transmit to others and can therefore be used as part of a risk reduction strategy. However, performance is lower in asymptomatic individuals than in symptomatic individuals and this limitation needs to be considered when designing testing programmes.
However, LFDs have generated considerable scientific and policy debate.3 They have imperfect sensitivity relative to PCR testing (ranging from <50% to >80%).4, 5, 6 Sensitivity is greater in symptomatic versus asymptomatic infection (eg, 72% vs 58%),4 which has led to concerns that LFDs might miss some infections and paradoxically increase transmission if individuals with false-negative results reduce transmission precautions.7 Conversely, LFDs need not detect and prevent all transmission to still have an effect at a population level; particularly where reproduction numbers are marginally greater than 1, an imperfect intervention might still be sufficient to control an outbreak.8, 9, 10, 11
An additional concern is the absence of a widely available proxy for an individual being infectious, as PCR positivity might persist for days to weeks after the end of symptoms or infectiousness.12, 13 Therefore, difficulties exist in directly assessing the sensitivity of LFDs in individuals who are infectious, which would be a better measure of their performance as a control intervention than sensitivity relative to PCR testing.14 The likelihood of onward transmission is also related to index patient viral load, with low measured PCR cycle threshold (Ct) values (a proxy for high viral loads in the tested individual) being associated with increased secondary cases.15, 16
LFDs are more sensitive as viral load increases;4, 16 therefore, test performance might be better understood with a curve reporting sensitivity at various viral loads, rather than by a single estimate that depends on the distribution of viral loads in the population studied. Combining index patient PCR viral loads, contact tracing data, and curves linking LFD performance with viral load, we previously estimated that LFDs could have detected 83–90% of infections leading to onward transmission in the SARS-CoV-2 alpha (B.1.1.7) variant period in the UK.16
In this study, we present data on the performance of LFDs relative to PCR across a range of settings during different variant waves of the pandemic. We evaluate if performance remained stable during the pre-alpha (B.1.177), alpha, delta (B.1.617.2), and omicron (BA.1 and BA.2) variant periods and after vaccinations. We combine our findings with contact tracing data to estimate the probable proportion of infectious individuals detected by LFDs with time.
Methods
Study design and participants
In this diagnostic study, we used LFD results paired with RT-PCR testing results, which had been prospectively collected to evaluate the performance of LFDs and their deployment, and for service quality assurance, by National Health Service (NHS) England's Test and Trace programme (now part of the UK Health Security Agency; UKHSA). Participants were asked to take a second test (LFD or RT-PCR) alongside their standard test (RT-PCR or LFD) specifically for the purposes of evaluating the LFDs and the testing programme. We analysed data collected from the start of the evaluation programme (Nov 4, 2020) until the end of provision of free testing for the general public (March 21, 2022). During this period, some participants might have contributed more than one set of paired samples. Details on each original study of the evaluation programme, including inclusion and exclusion criteria, study locations, participant consent, and recruitment (consecutive or random) are shown in the appendix (pp 10–13). Participants were predominantly 16 years or older, although some settings also included children aged 0–15 years. Further information on each study from the UK Government Department of Health and Social Care and UKHSA is available online.
National contact tracing data from England was obtained from NHS Test and Trace, as described previously,15, 16 for the period from Jan 1, 2021, to Jan 11, 2022, after which the national requirement for positive LFDs to be confirmed by RT-PCR was removed. Index cases were notified to the NHS Test and Trace service after a positive SARS-CoV-2 RT-PCR or LFD result in community-based or health-care-based settings. LFD results were self-reported to the service and RT-PCR results were reported by the testing centre. All index patients with a diagnostic RT-PCR test performed at three national testing laboratories (known as lighthouse laboratories) in Milton Keynes (UK), Alderley Park (Macclesfield, UK), and Glasgow (UK) were eligible for inclusion in this part of the study (appendix p 4). The three laboratories used the TaqPath COVID-19 assay (Thermo Fisher Scientific, Waltham, MA, USA) for testing.
Contacts, defined by NHS Test and Trace as people living in the same household as an index patient, or who had been at face-to-face distance from an index patient (within <1 m for ≥1 min or within <2 m for ≥15 min),16 were included in our analysis, provided they were only named by a single index patient in the 14 days either side of the index patient's positive test. Contacts with a positive RT-PCR or LFD result in the 1–10 days after the index patient's positive test were considered to represent plausible transmission events. The 1–10-day period was chosen to enrich for contacts for whom the index patient was the most likely source of SARS-CoV-2 infection.15, 16
The Research Ethics and Governance Group of the UKHSA (formerly Public Health England; London, UK) provided approval for the NHS Test and Trace evaluation programme studies. Approval was obtained for an umbrella framework and associated participant-facing materials for the prospective data collection elements of service evaluation and ongoing evaluation. The studies were reviewed and approved under the Research Ethics and Governance Group reference R&D 438 (appendix p 3).
Procedures
Evaluated LFDs were selected from those used in the UK's national testing programme. LFDs evaluated were the Innova SARS-CoV-2 Antigen Rapid Qualitative Test (Innova, in original packaging or repacked with individual buffer containers for the NHS; Xiamen Biotime Biotechnology, Fujian, China); the Orient Gene Rapid Covid-19 (Antigen) Self-Test (Zhejiang Orient Gene Biotech, Huzhou, China); and Acon Flowflex SARS-CoV-2 Antigen Rapid Test (Self-Testing; Acon Biotech [Hangzhou], Hangzhou, China). We refer to the test kits as Innova, Orient Gene, and Acon hereafter. RT-PCR testing was undertaken by routine laboratories within the NHS Test and Trace laboratory network, and was performed predominantly with the Thermo Fisher Scientific TaqPath COVID-19 assay. Additional details on the LFDs evaluated and RT-PCR tests are provided in the appendix (p 1). Thresholds for a positive RT-PCR test adopted by each laboratory matched those used for routine clinical reporting.
Paired LFD and RT-PCR testing was undertaken in several settings. Before wider national deployment, the performance of the Innova LFD was evaluated in two field studies (study ID numbers LFD001 and LFD002; appendix p 10), referred to as predeployment testing. All predeployment data in both studies were obtained from symptomatic testing sites, with a mixture of assisted and self-taken swabs. Subsequent settings included testing offered to asymptomatic individuals via unselected community-based testing (ie, the general public via testing sites and home testing; city-wide testing in Liverpool during a period of increased incidence17); targeted community testing (under-served groups identified by NHS Test and Trace as those likely to experience health inequalities and worse outcomes); schools (predominantly secondary); private and public sector workplaces; universities; and health-care workers. Paired testing was also undertaken for symptomatic individuals who attended local and regional symptomatic testing sites for RT-PCR testing (appendix pp 10–11).
Participants were provided with instructions from the NHS describing how to perform LFD testing and sampling for RT-PCR testing. A combined throat and nose or nose swab was obtained for each test. Details on swab types (combined nose and throat vs nose only) and sampling for LFDs and RT-PCR are provided in the appendix (p 1). RT-PCR and LFD swabs were self-taken (except in predeployment assisted testing) and instructed to be obtained within a few minutes of each other. Tests were denoted as assisted when the LFD result was interpreted by a trained individual. In self-testing, the user (or relevant person on behalf of the user such as a parent, guardian, or carer) interpreted the test themselves without assistance. Evaluations were done in live testing services; therefore, standard testing was always prioritised over the supplementary test, meaning swabbing order was not randomised during sample collection. If the standard testing was LFD, participants were asked to put the LFD to one side to develop, while providing the RT-PCR sample (so that they were unaware of the result of either test at sampling). Participants or operatives (where testing was assisted) were asked to interpret LFD results as positive, negative, or void, according to the manufacturer's instructions. Participants were unaware of RT-PCR results at the time of LFD testing. Similarly, laboratories undertaking RT-PCR testing were unaware of LFD results.
For LFD performance evaluations, the infecting variant in RT-PCR-positive infections was assigned on the basis of sequencing or genotyping when available and if not available, on the basis of the dominant circulating variant locally when the participant's home location was known, or otherwise nationally (appendix p 2). RT-PCR Ct values were used to estimate SARS-CoV-2 viral loads in swab fluids with use of calibrant samples (appendix p 14).
Participants provided information on symptoms. In accordance with national testing policy at the time, only participants reporting at least one of three cardinal symptoms (fever, dry continuous cough, or anosmia or ageusia) were considered symptomatic. Participants also self-reported their sex (male or female), date of birth, and vaccination status. Ethnicity data were not analysed.
Statistical analysis
All available data from the NHS Test and Trace studies were pooled in the current study, and thus no specific sample size calculation was used for the whole dataset (appendix p 2). Sample pairs with a void RT-PCR or LFD result were excluded from analysis. Similarly, sample pairs were excluded if the LFD manufacturer was not among the three evaluated, the PCR was performed with endpoint RT-PCR (which does not produce comparable estimates of viral load18) or an unknown PCR type, the RT-PCR was performed at a laboratory not participating in the study, or the participant withdrew from the study. Descriptive data are presented as proportions (n, %) or median (IQR), with sample numbers presented overall and according to test setting, RT-PCR positivity, and assigned variant.
RT-PCR-positive samples were used to analyse LFD sensitivity. We used univariable and multivariable logistic regression to model relationships between LFD positivity and log10 viral load and other available covariates. We accounted for any non-linearity of associations with continuous variables using natural cubic splines where these improved model fit according to the Bayesian information criterion (appendix p 2). In the regression analyses, we modelled the odds of a positive LFD result. The included covariates were LFD manufacturer, test setting, assisted testing versus self-testing, age (as a continuous variable), self-reported sex, symptom status (symptomatic; ie, any of fever, cough, or anosmia or ageusia; otherwise asymptomatic), vaccination status (unvaccinated, one dose, two or more doses, or unknown), and assigned viral variant (alpha or pre-alpha; delta; omicron; or other or unknown).19
We used RT-PCR-negative samples to analyse LFD specificity using univariable and multivariable logistic regression and the same covariates. In these analyses, we modelled the odds of a false-positive test, such that higher odds ratios (ORs) indicated lower specificity. Locally dominant circulating variants, or nationally dominant variants when a participant's home location was not recorded, were included in specificity analyses to capture temporal changes that occurred alongside changes in circulating variants, as it was not feasible to also include calendar time in all models due to collinearity with circulating variants. Sample pairs with missing covariate data were excluded from regression analyses.
We used contact tracing data and logistic regression to estimate the relationship between index patient symptom status and RT-PCR Ct value (viral load), and positive results in RT-PCR-tested or LFD-tested contacts, adjusting for multiple index patient and contact factors (appendix pp 2–3). Following a similar approach to previous analyses,15, 16 we used case–contact pairs plausibly related by transmission to estimate the proportion of infectious index patients potentially detected by LFDs. We applied the estimated performance of the most widely used LFD (Innova) in unselected community-based testing to contact tracing data from all index patients with at least one RT-PCR-positive or LFD-positive contact to determine overall estimated proportion of potentially detectable index patients. Estimates of infectious patients detected by LFDs accounted for symptom status of the index patient and index patient viral load at diagnosis. Results were visualised by index patient test month, and plotted according to the nationally dominant circulating variant at index patient diagnosis (alpha, Jan 1, 2021, to May 18, 2021; delta, May 19 to Dec 12, 2021; and omicron, Dec 13, 2021, to Jan 11, 2022). We used these combined estimates of LFD performance, together with varying assumptions for the prevalence of SARS-CoV-2 infection and symptoms from other causes, to calculate the number of symptomatic and asymptomatic individuals needed to be tested to detect one infection that would otherwise go on to transmit the virus.
All analyses were performed with R (version 4.2). Findings are summarised with point estimates, 95% CIs, and p values without threshold-based interpretations of statistical significance in accordance with the American Statistical Society20 and other reporting best practice for observational studies.
Role of the funding source
LFD evaluations were designed, and data collection commissioned, by the UKHSA and UK Government Department of Health and Social Care. Data were analysed and interpreted independently at the University of Oxford and UKHSA to confirm results. Authors who are employees of the UKHSA had a role in writing the report.
Results
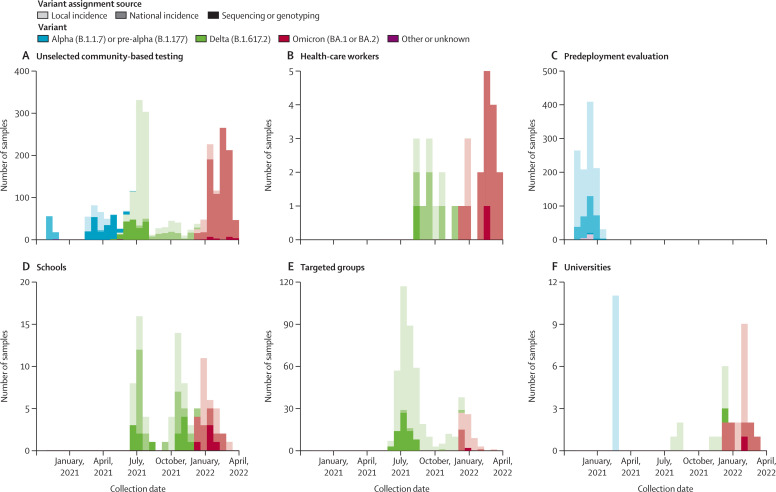
Between Nov 4, 2020, and March 21, 2022, 83 280 paired LFD and RT-PCR tests were performed. 7898 paired tests were excluded: 23 LFD tests were performed with an LFD manufacturer other than the three evaluated, 6389 PCR tests were tested with endpoint RT-PCR or an unknown PCR type, 46 RT-PCR tests were done at laboratories not participating in the study, 1272 RT-PCR tests had a void result, 147 LFD tests had a void result, and for 21 sample pairs the participants withdrew consent (appendix p 5). Thus, 75 382 paired tests were eligible for analysis. Of these, 4131 (5·5%) samples were RT-PCR-positive. The study set comprised 26 797 samples from male participants, 30 940 samples from female participants, and 17 645 samples from participants for whom sex was not recorded. The median age was 39 (IQR 27–53) years. The test settings were predeployment evaluation sites (1123 [16·6%] RT-PCR-positive samples of 6759 total samples), unselected community-based testing (2381 [7·4%] of 32 266), health-care workers (28 [1·8%] of 1587), schools (89 [0·7%] of 12 397), workplaces (seven [0·1%] of 5700), universities (38 [0·6%] of 6456), and specifically targeted under-served groups (465 [4·6%] of 10 217; figure 1 , appendix p 6).
Figure 1.
SARS-CoV-2 RT-PCR-positive samples used in lateral flow device evaluation by setting and viral variant
The source of the assigned SARS-CoV-2 variant is also shown by colour gradient. Seven RT-PCR-positive results from workplaces, obtained between April 27 and Sept 1, 2021, are not shown.
The overall sensitivity of LFDs relative to RT-PCR-testing was 63·2% (95% CI 61·7–64·6; 2609 LFD-positive samples of 4131 RT-PCR-positive samples). Sensitivity was 71·6% (69·8–73·4) in unselected community-based testing, which was higher than in predeployment testing (52·8% [49·8–55·8]; table 1 ). Sensitivity was 55·7% (53·1–58·2) when the assigned variant was alpha or pre-alpha, 64·0% (61·6–66·4) for delta, and 73·0% (70·2–75·6) for omicron. Sensitivity was higher in participants who had received one vaccine dose (67·6% [63·3–71·7]) or two or more vaccine doses (69·7% [67·3–72·0]) versus unvaccinated participants (57·3% [55·1–59·4]). Sensitivity was higher in symptomatic participants (68·7% [66·9–70·4]) than in asymptomatic participants (52·8% [50·1–55·4]). Sensitivity was higher for the Acon LFD (72·3% [66·4–77·8]) and Orient Gene LFD (69·7% [65·8–73·3]) than with the Innova LFD (61·3% [59·6–62·9]).
Table 1.
LFD performance in SARS-CoV-2 PCR-positive samples (n=4131)
|
Summary |
Univariable |
Multivariable |
||||||
|---|---|---|---|---|---|---|---|---|
| LFD negative; n (%) or median (IQR); n=1522 | LFD positive; n (%) or median (IQR); n=2609 | Sensitivity (95% CI) | OR (95% CI) | p value | Adjusted OR (95% CI) | p value | ||
| Log10 viral load* | 3·84 (2·48–5·02) | 6·12 (5·29–6·80) | .. | 2·80 (2·63–2·98) | <0·0001 | 2·85 (2·66–3·06) | <0·0001 | |
| LFD type | ||||||||
| Innova | 1270 (38·7%) | 2008 (61·3%) | 61·3% (59·6–62·9) | 1 (ref) | .. | 1 (ref) | .. | |
| Acon | 70 (27·7%) | 183 (72·3%) | 72·3% (66·4–77·8) | 1·65 (1·24–2·20) | 0·0005 | 1·65 (0·96–2·83) | 0·070 | |
| Orient Gene | 182 (30·3%) | 418 (69·7%) | 69·7% (65·8–73·3) | 1·45 (1·20–1·75) | 0·0001 | 1·11 (0·78–1·57) | 0·56 | |
| Test setting | ||||||||
| Unselected community-based testing | 676 (28·4%) | 1705 (71·6%) | 71·6% (69·8–73·4) | 1 (ref) | .. | 1 (ref) | .. | |
| Health-care workers | 10 (35·7%) | 18 (64·3%) | 64·3% (44·1–81·4) | 0·71 (0·33–1·55) | 0·40 | 0·60 (0·14–2·50) | 0·48 | |
| Predeployment evaluation | 530 (47·2%) | 593 (52·8%) | 52·8% (49·8–55·8) | 0·44 (0·38–0·51) | <0·0001 | 0·51 (0·36–0·73) | 0·0002 | |
| Schools | 52 (58·4%) | 37 (41·6%) | 41·6% (31·2–52·5) | 0·28 (0·18–0·43) | <0·0001 | 0·70 (0·30–1·63) | 0·41 | |
| Targeted groups | 233 (50·1%) | 232 (49·9%) | 49·9% (45·3–54·5) | 0·39 (0·32–0·48) | <0·0001 | 0·51 (0·33–0·78) | 0·0018 | |
| Universities | 16 (42·1%) | 22 (57·9%) | 57·9% (40·8–73·7) | 0·55 (0·28–1·04) | 0·067 | 0·84 (0·31–2·33) | 0·74 | |
| Workplaces | 5 (71·4%) | 2 (28·6%) | 28·6% (3·7–71·0) | 0·16 (0·03–0·82) | 0·028 | 0·12 (0·01–1·86) | 0·13 | |
| Age† | 34 (25–47) | 36 (25–47) | .. | 1·00 (1·00–1·01) | 0·34 | 1·00 (0·99–1·00) | 0·41 | |
| Unknown‡ | 105 | 151 | NA | .. | .. | .. | .. | |
| Self-reported sex | ||||||||
| Female | 740 (35·9%) | 1323 (64·1%) | 64·1% (62·0–66·2) | 1 (ref) | .. | 1 (ref) | .. | |
| Male | 680 (37·4%) | 1139 (62·6%) | 62·6% (60·3–64·8) | 0·94 (0·82–1·07) | 0·33 | 0·89 (0·75–1·07) | 0·21 | |
| Unknown‡ | 102 | 147 | NA | .. | .. | .. | .. | |
| Assistance in interpreting test result | ||||||||
| Assisted | 602 (47·5%) | 665 (52·5%) | 52·5% (49·7–55·3) | 1 (ref) | .. | 1 (ref) | .. | |
| Self-interpreted | 920 (32·1%) | 1944 (67·9%) | 67·9% (66·1–69·6) | 1·91 (1·67–2·19) | <0·0001 | 0·79 (0·60–1·05) | 0·11 | |
| Symptom status | ||||||||
| Asymptomatic | 648 (47·2%) | 724 (52·8%) | 52·8% (50·1–55·4) | 1 (ref) | .. | 1 (ref) | .. | |
| Symptomatic | 847 (31·3%) | 1859 (68·7%) | 68·7% (66·9–70·4) | 1·96 (1·72–2·25) | <0·0001 | 1·63 (1·30–2·04) | <0·0001 | |
| Unknown‡ | 27 | 26 | NA | .. | .. | .. | .. | |
| Vaccination status | ||||||||
| Unvaccinated | 874 (42·7%) | 1171 (57·3%) | 57·3% (55·1–59·4) | 1 (ref) | .. | 1 (ref) | .. | |
| One dose | 163 (32·4%) | 340 (67·6%) | 67·6% (63·3–71·7) | 1·56 (1·27–1·91) | <0·0001 | 1·13 (0·81–1·57) | 0·48 | |
| Two or more doses | 471 (30·3%) | 1083 (69·7%) | 69·7% (67·3–72·0) | 1·72 (1·49–1·97) | <0·0001 | 1·13 (0·81–1·59) | 0·46 | |
| Unknown‡ | 14 | 15 | NA | .. | .. | .. | .. | |
| Assigned SARS-CoV-2 variant | ||||||||
| Alpha (B.1.1.7) or pre-alpha (B.1.177) | 674 (44·3%) | 846 (55·7%) | 55·7% (53·1–58·2) | 1 (ref) | .. | 1 (ref) | .. | |
| Delta (B.1.617.2) | 545 (36·0%) | 970 (64·0%) | 64·0% (61·6–66·4) | 1·42 (1·23–1·64) | <0·0001 | 1·00 (0·69–1·45) | 0·99 | |
| Omicron (BA.1 or BA.2) | 290 (27·0%) | 783 (73·0%) | 73·0% (70·2–75·6) | 2·15 (1·82–2·55) | <0·0001 | 1·63 (1·02–2·59) | 0·042 | |
| Other or unknown | 13 (56·5%) | 10 (43·5%) | 43·5% (23·2–65·5) | 0·61 (0·27–1·41) | 0·25 | 0·43 (0·15–1·23) | 0·12 | |
Denominators for percentages are the total number of LFD-negative and LFD-positive samples in each row. The logistic regression analyses show ORs for LFD positivity, such that higher ORs represent greater sensitivity. There was no evidence that adjusted models allowing for a non-linear relationship between the natural log odds of a positive result and log10 viral load or age (using splines with up to five knots) improved model fit based on the Bayesian information criterion. LFD=lateral flow device. n=number of samples. OR=odds ratio. NA=not applicable.
OR is for the odds of LFD positivity per 1 log10 copies per mL increase in viral load.
ORs per 1-year increase.
Data not summarised or analysed as there is no obvious interpretation.
After adjustment for covariates, increased viral load was independently associated with a positive LFD result (per 1 log10 copies per mL increase, adjusted OR [aOR] 2·85 [95% CI 2·66–3·06]; p<0·0001; table 1). For instance, the Innova LFD had predicted sensitivity for omicron infection in symptomatic participants in unselected community-based settings of 28·9% (20·8–38·7) at a viral load of 103 copies per mL, 77·6% (69·6–83·9) at a viral load of 105 copies per mL, and 96·7% (95·0–97·9) at a viral load of 107 copies per mL. Corresponding values for asymptomatic testing in unselected community settings were 19·9% (13·6–28·2), 67·9% (57·9–76·5), and 94·7% (91·9–96·6). Although estimates were compatible with no important difference in sensitivity between the Acon and Innova LFDs, we found that the Acon LFD might have been more sensitive than the Innova LFD (aOR 1·65 [95% CI 0·96–2·83]; p=0·070). There was no evidence of a difference between Innova and Orient Gene LFDs (aOR 1·11 [0·78–1·57]; p=0·56; table 1, figure 2 , appendix p 7).
Figure 2.
Sensitivity of SARS-CoV-2 LFDs by viral load and patient symptoms
The model predictions plotted are adjusted for test setting (predictions are shown for unselected community-based testing), assistance performing the test (self-performed), vaccination status (unvaccinated), and variant (alpha [B.1.1.7] or pre-alpha [B1.1.177]). In addition to the model shown in table 1, an interaction term between viral load and LFD was included to allow the shape of the curves to vary by device. A comparison with observed data is presented in the appendix (p 7). LFD=lateral flow device.
Compared with unselected community-based testing, sensitivity was independently lower in predeployment testing (aOR 0·51 [95% CI 0·36–0·73]; p=0·0002) and in targeted groups (aOR 0·51 [0·33–0·78]; p=0·0018; table 1). Symptomatic participants were independently more likely to test positive than asymptomatic participants (aOR 1·63 [1·30–2·04]; p<0·0001), while there was no evidence of a difference in LFD sensitivity with or without assistance in interpreting the test. Additionally, vaccination status, age, and sex did not show independent associations with LFD results. LFD sensitivity did not differ with delta infections compared with alpha or pre-alpha infections (aOR 1·00 [0·69–1·45]; p=0·99), but omicron infections were more likely to be LFD positive than alpha or pre-alpha infections (aOR 1·63 [1·02–2·59]; p=0·042).
The overall specificity of LFDs was 99·71% (95% CI 99·66–99·74; 71 041 LFD-negative samples of 71 251 RT-PCR-negative samples) and was greater than 99·4% across all settings (table 2 ). In multivariable modelling, compared with the Innova LFD, Acon devices were associated with more false-positive results (aOR 2·00 [95% CI 1·21–3·29]; p=0·0065), with weak evidence that Orient Gene devices might have led to fewer false-positive results (aOR 0·49 [0·23–1·04]; p=0·065; table 2). There were fewer false-positive results in schools than in unselected community-based testing (aOR 0·33 [0·16–0·68]; 0·0025), whereas false-positive results were more common in predeployment testing (aOR 2·09 [1·15–3·82]; p=0·016). Self-interpreted tests resulted in more false-positives than assisted tests (aOR 2·61 [1·46–4·67]; p=0·0012). False-positive results were more common in symptomatic participants than in asymptomatic participants (aOR 2·15 [1·48–3·13]; p=0·0001). Specificity did not differ by vaccination status, age, or sex. Based on locally or nationally dominant circulating variants, false-positive tests were more common during the delta period (aOR 2·68 [1·43–5·00]; p=0·0020) and omicron period (aOR 5·47 [2·54–11·81]; p<0·0001) than during the alpha and pre-alpha period.
Table 2.
LFD performance in SARS-CoV-2 PCR-negative samples (n=71 251)
|
Summary |
Univariable |
Multivariable |
||||||
|---|---|---|---|---|---|---|---|---|
| LFD positive; n (%); n=210 | LFD negative; n (%); n=71 041 | Specificity (95% CI) | OR (95% CI) | p value | Adjusted OR (95% CI) | p value | ||
| LFD type | ||||||||
| Innova | 156 (0·3%) | 59 884 (99·7%) | 99·7% (99·7–99·8) | 1 (ref) | .. | 1 (ref) | .. | |
| Acon | 43 (0·7%) | 5712 (99·3%) | 99·3% (99·0–99·5) | 2·62 (1·86–3·67) | <0·0001 | 2·00 (1·21–3·29) | 0·0065 | |
| Orient Gene | 11 (0·2%) | 5445 (99·8%) | 99·8% (99·6–99·9) | 0·70 (0·38–1·29) | 0·26 | 0·49 (0·23–1·04) | 0·065 | |
| Test setting | ||||||||
| Unselected community-based testing | 130 (0·4%) | 29 755 (99·6%) | 99·6% (99·5–99·6) | 1 (ref) | .. | 1 (ref) | .. | |
| Health-care workers | 7 (0·4%) | 1552 (99·6%) | 99·6% (99·1–99·8) | 1·03 (0·48–2·21) | 0·93 | 1·33 (0·48–3·71) | 0·59 | |
| Predeployment evaluation | 34 (0·6%) | 5602 (99·4%) | 99·4% (99·2–99·6) | 1·39 (0·95–2·03) | 0·089 | 2·09 (1·15–3·82) | 0·016 | |
| Schools | 16 (0·1%) | 12 292 (99·9%) | 99·9% (99·8–99·9) | 0·30 (0·18–0·50) | <0·0001 | 0·33 (0·16–0·68) | 0·0025 | |
| Targeted groups | 20 (0·2%) | 9732 (99·8%) | 99·8% (99·7–99·9) | 0·47 (0·29–0·75) | 0·0017 | 0·97 (0·45–2·09) | 0·94 | |
| Universities | 3 (<0·1%) | 6415 (>99·9%) | >99·9% (99·9–100·0) | 0·11 (0·03–0·34) | 0·0001 | 0·34 (0·10–1·16) | 0·085 | |
| Workplaces* | 0 | 5693 (100%) | 100·0% (99·9–100·0) | NA | .. | NA | .. | |
| Assistance in interpreting test result | ||||||||
| Assisted | 41 (0·1%) | 30 095 (99·9%) | 99·9% (99·8–99·9) | 1 (ref) | .. | 1 (ref) | .. | |
| Self-interpreted | 169 (0·4%) | 40 946 (99·6%) | 99·6% (99·5–99·6) | 2·46 (1·75–3·46) | <0·0001 | 2·61 (1·46–4·67) | 0·0012 | |
| Symptom status | ||||||||
| Asymptomatic | 119 (0·2%) | 56 766 (99·8%) | 99·8% (99·7–99·8) | 1 (ref) | .. | 1 (ref) | .. | |
| Symptomatic | 87 (0·8%) | 10 640 (99·2%) | 99·2% (99·0–99·3) | 3·65 (2·77–4·82) | <0·0001 | 2·15 (1·48–3·13) | 0·0001 | |
| Unknown† | 4 | 3635 | NA | .. | .. | .. | .. | |
| Vaccination status | ||||||||
| Unvaccinated | 73 (0·3%) | 24 677 (99·7%) | 99·7% (99·6–99·8) | 1 (ref) | .. | 1 (ref) | .. | |
| One dose | 27 (0·2%) | 12 263 (99·8%) | 99·8% (99·7–99·9) | 0·89 (0·57–1·39) | 0·61 | 0·76 (0·43–1·35) | 0·35 | |
| Two or more doses | 110 (0·3%) | 32 681 (99·7%) | 99·7% (99·6–99·7) | 1·08 (0·81–1·46) | 0·60 | 0·81 (0·47–1·40) | 0·44 | |
| Unknown† | 0 | 1420 | NA | .. | .. | .. | .. | |
| Dominant circulating SARS-CoV-2 variant | ||||||||
| Alpha (B.1.1.7) or pre-alpha (B.1.177) | 53 (0·2%) | 24 440 (99·8%) | 99·8% (99·7–99·8) | 1 (ref) | .. | 1 (ref) | .. | |
| Delta (B.1.617.2) | 114 (0·3%) | 39 297 (99·7%) | 99·7% (99·7–99·8) | 1·16 (0·84–1·61) | 0·38 | 2·68 (1·43–5·00) | 0·0020 | |
| Omicron (BA.1 or BA.2) | 41 (0·6%) | 7098 (99·4%) | 99·4% (99·2–99·6) | 2·21 (1·47–3·33) | 0·0001 | 5·47 (2·54–11·81) | <0·0001 | |
| Other or unknown | 2 (1·0%) | 206 (99·0%) | 99·0% (96·6–99·9) | 3·78 (0·91–15·60) | 0·066 | 2·52 (0·58–10·93) | 0·22 | |
| Age‡ | 36 (27–49) | 39 (27–53) | .. | 0·99 (0·98–1·00) | 0·11 | 0·99 (0·98–1·00) | 0·20 | |
| Unknown† | 25 | 17 194 | NA | .. | .. | .. | .. | |
| Self-reported sex | ||||||||
| Female | 102 (0·4%) | 28 775 (99·6%) | 99·6% (99·6–99·7) | 1 (ref) | 1 (ref) | |||
| Male | 83 (0·3%) | 24 895 (99·7%) | 99·7% (99·6–99·7) | 0·99 (0·74–1·33) | 0·96 | 1·06 (0·78–1·43) | 0·72 | |
| Unknown† | 25 | 17 371 | NA | .. | .. | .. | .. | |
Denominators for percentages are the total number of LFD-positive and LFD-negative samples in each row. The logistic regression analyses show ORs for a false-positive test (ie, higher ORs indicate lower specificity). There was no evidence that adjusted models allowing for a non-linear relationship between the natural log odds of a false-positive result and age (using splines with up to five knots) improved model fit based on the Bayesian information criterion. LFD=lateral flow device. n=number of samples. OR=odds ratio. NA=not applicable.
No false-positive tests in workplace testing and so results were excluded from regression models.
Data not summarised or analysed as there is no obvious interpretation.
ORs per 1-year increase.
We estimated the proportion of infectious individuals potentially detectable by LFDs on the basis of Innova LFD performance in unselected community-based testing. Between Jan 1, 2021, and Jan 11, 2022, 6 263 786 contacts of RT-PCR-positive SARS-CoV-2 index patients were identified. Of these contacts, 1 173 643 (18·7%) underwent an RT-PCR test or self-reported doing an LFD test within 1–10 days after the index patient's RT-PCR test, and 377 151 (6·0%) tested positive for SARS-CoV-2. Contacts who tested positive were linked to 347 374 unique index patients, with 322 416 (92·8%) of these index patients linked to a single SARS-CoV-2-positive contact, 21 321 (6·1%) to two positive contacts, and 3637 (1·0%) to three or more positive contacts (appendix p 8).
We found that tested contacts of index patients with high viral loads (low Ct values) were more likely to test positive than the contacts of index patients with low viral loads. In the same regression analysis, we found that the contacts of symptomatic index patients were more likely to test positive than those of asymptomatic index patients at a given viral load (figure 3A ). Additionally, viral loads in asymptomatic index patients were lower than in symptomatic index patients throughout the alpha, delta, and omicron periods (appendix p 9).
Figure 3.
Index patient Ct values and the probability of tested contacts being RT-PCR positive or LFD positive (A) and estimated index cases detectable by LFDs among case–contact pairs with probable transmission (B)
Contacts of asymptomatic index patients were 0·76 times as likely to test positive as contacts of symptomatic index patients at a Ct value of 10; 0·70 times as likely at a Ct value of 20; and 0·65 times as likely at a Ct value of 30. The model in part A was adjusted for index patient age (predictions plotted for age 40 years), index patient sex (female), index patient vaccination status (boosted, ≥3 vaccine doses), contact event type (household or accommodation), contact age (40 years), contact sex (female), and index test date (July 1, 2021). We found no evidence that fitting an interaction between index patient symptom status and Ct values improved model fit. In part B, error bars indicate 95% CIs calculated by non-parametric bootstrapping (1000 iterations). Ct=cycle threshold. LFD=lateral flow device.
78·3% (95% CI 75·3–81·2) of index patients with one or more RT-PCR-positive or LFD-positive contacts (ie, representing plausible onward transmission) were estimated to have been detectable by the Innova LFD. Similar detection rates were estimated in index patients with one (78·3% [75·3–81·1]), two (79·1% [76·2–82·0]), or three or more (78·5% [75·3–81·6]) SARS-CoV-2-positive contacts. Among the 347 374 index patients with one or more RT-PCR-positive or LFD-positive contacts, 20 712 (6·0%) were recorded as asymptomatic and 326 662 (94·0%) as symptomatic. The estimated proportion of index patients detectable by Innova LFD was higher with symptoms (79·7% [76·7–82·5]) than without (57·6% [53·6–61·9]). Applying estimates from this study, the number of LFD tests among symptomatic individuals that would be needed to detect one case that would otherwise go on to transmit virus ranged from 13 to 232 under varying scenarios for the prevalence of SARS-CoV-2 and symptoms from other sources. Under the same conditions, the number of asymptomatic tests needed to prevent one transmission was more than 1000 except in the scenario of perfect LFD performance (appendix p 15).
The estimated proportion of index patients detectable by Innova LFD was similar with time and during the alpha, delta, and omicron periods for symptomatic patients (figure 3B, table 3 ). For asymptomatic index patients, the estimated proportion of index patients detectable by LFDs was consistently lower than for symptomatic index patients with time and across variants, although 95% CIs overlapped in some time intervals. Among asymptomatic index patients, the estimated proportion detectable by LFDs was higher during the omicron period than during the alpha and delta periods although 95% CIs overlapped. The increase during the omicron period is likely to reflect the increase in measured sensitivity of LFDs during omicron circulation rather than a change in viral load distribution (appendix p 9).
Table 3.
Estimated index cases detectable by LFDs by dominant circulating variant and symptom status among case–contact pairs with probable SARS-CoV-2 transmission
| Index case symptom status | Number of index cases | Estimated percentage of index cases detectable by LFDs (95% CI) | |
|---|---|---|---|
| Alpha | Symptomatic | 60 322 | 76·5% (69·2–85·5) |
| Delta | Symptomatic | 198 707 | 80·5% (73·7–86·9) |
| Omicron | Symptomatic | 67 633 | 80·1% (74·6–85·3) |
| Alpha | Asymptomatic | 3590 | 51·1% (15·3–67·0) |
| Delta | Asymptomatic | 13 153 | 57·2% (31·3–80·8) |
| Omicron | Asymptomatic | 3969 | 64·9% (54·5–80·1) |
Pango lineages: alpha (B.1.1.7), delta (B.1.617.2), and omicron (BA.1 or BA.2). LFD=lateral flow device.
Discussion
In a national LFD evaluation programme, the sensitivity of LFDs compared with RT-PCR was 71·6% (95% CI 69·8–73·4) in unselected community-based testing. In a multivariable analysis, sensitivity was similar during the periods of estimated alpha and pre-alpha infections and delta infections, and increased during the omicron period (relative to the alpha and pre-alpha period), potentially reflecting changes in the virus or testing proficiency and human behaviour. This finding is in contrast with previous reports of lower sensitivity with omicron versus delta infections,21, 22 and others have reported similar performance with omicron versus delta.23 Conversely, although specificity remained higher than 99%, it decreased as successive variants dominated, reflecting changes with time in testing practice or another factor we did not adjust for, rather than in the variant itself, as these samples were RT-PCR negative. We found no evidence that LFD sensitivity or specificity was independently associated with vaccination status, age, or sex after adjusting for viral load and other factors. Vaccination has been reported to reduce viral load in alpha breakthrough infections, but not in delta infections.15
Symptomatic individuals had increased viral loads; even adjusting for viral load, having symptoms (fever, cough, or anosmia or ageusia) was associated with increased LFD sensitivity. This finding might reflect different antigen to RNA ratios with different symptoms, although we did not investigate if LFD performance varied with specific individual symptoms. With a one-off screen, asymptomatic infections might be detected later when residual RNA might be present with less antigen, compared with early infection when most symptomatic patients are prompted to test by the onset of symptoms.
The Acon LFD might have been more sensitive than Innova LFD, particularly at lower viral loads, but also had the lowest specificity, reflecting trade-offs in how devices are calibrated. We found no evidence of a difference between Innova and Orient Gene sensitivity in terms of adjusted odds of LFD positivity; however, power was limited as most tests used Innova LFDs. Sensitivity with Acon and Orient Gene was achieved with nasal swabs only, whereas Innova requires combined nose and throat swabs. Regarding overall functionality of the LFDs, there were fewer reported void LFD results than void RT-PCR results.
Similar to previous studies,13, 24, 25, 26 we found that participants with high viral loads (ie, those who were more infectious) were more likely to be detected by LFDs than those with lower viral loads. Among case–contact pairs with plausible transmission, the proportion of index patients detectable by community-based Innova LFD testing was 78·3%, similar to previous estimates for this device (83·0% [95% CI 82·8–83·1]).16 This estimate is higher than the overall sensitivity of LFDs (71·6%) in unselected community-based testing. However, LFDs were estimated to detect fewer asymptomatic index patients than symptomatic index patients (57·6% vs 79·7%). This result was due to both reduced sensitivity at a given viral load and reduced viral loads in asymptomatic infections. Offsetting this outcome, we found that asymptomatic individuals were less infectious than symptomatic individuals at a given viral load; and the current and previous findings indicate an association of low viral loads in asymptomatic infections with reduced transmission.15 Notably, asymptomatic index patients are also likely to be under-ascertained in national contact tracing data, especially during periods when asymptomatic screening was uncommon.
Our findings support the use of LFDs as a mechanism to detect potentially infectious individuals and reduce transmission, particularly as these tests perform best in individuals most likely to be infectious (ie, symptomatic or high viral load infections). However, we show, alongside other studies, that LFD performance in asymptomatic individuals might be lower than is generally understood.4, 25, 27 LFDs have been deployed in several settings, including in population-wide mass testing of asymptomatic individuals. In this study, a combination of population prevalence, lower viral loads in asymptomatic individuals, and lower onward transmission at a given viral load compared with symptomatic patients means that even with perfect LFD sensitivity, potentially several thousand asymptomatic people would need to be tested to detect one patient who would otherwise transmit the virus onwards (appendix p 15). With imperfect LFD sensitivity, more individuals would need to be tested to detect one potentially transmitting patient. If false-negative results do not change behaviour, then reduced LFD sensitivity acts simply to make testing programmes less efficient and more costly per transmission averted, but still potentially effective at reducing the spread of infections. Thus, if people plan to participate in activities when they do not have access to an LFD, providing access to LFDs might act to reduce the risk of transmission, even if imperfectly. Risk reduction might also be important when the consequences of transmission are high and those tested are likely to maintain transmission precautions with a negative result (eg, in health care or social care). However, some caution is required if LFDs are used by individuals to relax adhering to transmission precautions,28, 29 in which case false-negative results with a poorly sensitive test could lead to additional transmission. These factors need to be considered when assessing implementation and messaging around asymptomatic screening programmes. Potential personal or societal effects of false-positive tests and apparently small differences in specificity by device also need to be considered even for devices with high specificity as in the current study (>99%), as many thousands of tests might be done.
Our evaluation of LFD performance has several limitations. RT-PCR is an imperfect reference standard given the persistence of PCR positivity for days to weeks after infection.12, 13 False-negative RT-PCR tests could also lead to underestimation of specificity. Primary diagnostic tests were always performed first, rather than randomising the order of swabs taken for LFDs and RT-PCR, however the effect of this is uncertain. Variations between laboratories in RT-PCR assays and thresholds for positive results are an additional possible source of variation; however, calibrants were used to convert Ct values across assays to estimated viral loads in common units. Our calibration approach was widely deployable, however more accurate approaches for quantifying viral load including droplet digital PCR exist. Additionally, estimated viral loads in samples only approximate the actual viral loads in participants, given that respiratory samples are based on swabs rather than direct body fluid sampling. Although we show that asymptomatic individuals were less likely to be detected by LFDs, in part this might reflect the performance of a single test capturing some infections several days after they started. Regular asymptomatic testing could have improved performance, as this might detect incident infections earlier in the course of infection. Digital LFD readers were not used, which might increase accuracy, as in national evaluations in the UK.30, 31 Although numerically the incidence of SARS-CoV-2 infection does not change sensitivity or specificity, changes in incidence could affect testing behaviour and viral load distributions. A further variable not studied in isolation was the effect of time; the close association of calendar time with variants and the vaccine programme meant that we could not analyse the effect of time separately.
Our transmission analysis also has limitations. Identification of case–contact pairs relied on both index patients and contacts participating in SARS-CoV-2 testing, with demographic, socioeconomic, and behavioural factors affecting test-seeking. During the study, RT-PCR testing was provided for symptomatic individuals or after a positive LFD test. However, some asymptomatic individuals also sought PCR testing for other reasons (eg, after contact events). Therefore, case–contact pairs involving an asymptomatic patient might have been enriched for pairs with contact with a third party who was the true source of both infections. In this scenario, the properties of the asymptomatic infection are not what determined transmission.
In summary, LFDs have remained able to detect most SARS-CoV-2 infections throughout the roll-out of vaccination and across different viral variants. LFDs were estimated to detect most infections that have the potential to transmit to others; however, LFD performance was lower in asymptomatic individuals than in symptomatic individuals, and this needs to be considered when designing testing programmes. Ongoing monitoring of performance with new variants is also required.
For the UK Government Department of Health and Social Care and UKHSA data see https://www.gov.uk/government/publications/lateral-flow-device-performance-data.
Data sharing
Data collected for the study, including de-identified individual participant data and a data dictionary defining each field in the set, will be available from NHS Digital's Data Access Request Service with publication of this Article. Details of how to apply for access to the data via the Data Access Request Service are provided at https://digital.nhs.uk/services/data-access-request-service-dars.
Acknowledgments
Acknowledgments
This study was funded by the UK Health Security Agency (UKHSA; formerly Public Health England); the UK Government Department of Health and Social Care; the National Institute for Health Research (NIHR) Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance, University of Oxford, in partnership with UKHSA; and the University of Oxford NIHR Biomedical Research Centre. DWE is a Robertson Foundation Fellow.
Contributors
DWE, MF, TEAP, and TF conceived the study. DWE, MF, ST, JC-H, RS, NG, ARD, PEK, MS, CK, PM, and EB contributed to the investigation. DWE and MF did the formal analysis and directly accessed and verified the underlying data reported. SH, TEAP, and TF supervised the study. DWE and MF were responsible for data visualisation. DWE, MF, and JW wrote the original manuscript draft. All authors reviewed and edited the manuscript and had full access to all the data in the study. All authors had final responsibility for the decision to submit for publication.
Declaration of interests
DWE has received lecture fees from Gilead Sciences outside the submitted work. All other authors declare no competing interests.
Supplementary Material
References
- 1.Mahase E. Covid-19: UK regulator approves lateral flow test for home use despite accuracy concerns. BMJ. 2020;371 doi: 10.1136/bmj.m4950. [DOI] [PubMed] [Google Scholar]
- 2.NHS England NHS England and NHS Improvement rollout of lateral flow devices for asymptomatic staff testing for SARS CoV-2 (phase 2: trusts) Nov 16, 2020. https://www.england.nhs.uk/coronavirus/documents/nhs-england-and-nhs-improvement-rollout-of-lateral-flow-devices-for-asymptomatic-staff-testing-for-sars-cov-2-phase-2-trusts/
- 3.Deeks J, Raffle A, Gill M. Covid-19: government must urgently rethink lateral flow test roll out. Jan 12, 2023. https://blogs.bmj.com/bmj/2021/01/12/covid-19-government-must-urgently-rethink-lateral-flow-test-roll-out/
- 4.Dinnes J, Sharma P, Berhane S, et al. Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2022;7 doi: 10.1002/14651858.CD013705.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mistry DA, Wang JY, Moeser M-E, Starkey T, Lee LYW. A systematic review of the sensitivity and specificity of lateral flow devices in the detection of SARS-CoV-2. BMC Infect Dis. 2021;21:828. doi: 10.1186/s12879-021-06528-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.UK Government Department of Health and Social Care Asymptomatic testing for SARS-CoV-2 using antigen-detecting lateral flow devices. July 7, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/999866/asymptomatic-testing-for-SARS-CoV-2-using-antigen-detecting-lateral-flow-devices-evidence-from-performance-data-Oct-2020-to-May-2021.pdf
- 7.Deeks JJ, Raffle AE. Lateral flow tests cannot rule out SARS-CoV-2 infection. BMJ. 2020;371 doi: 10.1136/bmj.m4787. [DOI] [PubMed] [Google Scholar]
- 8.Howard J, Huang A, Li Z, et al. An evidence review of face masks against COVID-19. Proc Natl Acad Sci U S A. 2021;118:e201. doi: 10.1073/pnas.2014564118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris DH, Rossine FW, Plotkin JB, Levin SA. Optimal, near-optimal, and robust epidemic control. Commun Phys. 2021;4:78. [Google Scholar]
- 10.Fisman DN, Greer AL, Tuite AR. Bidirectional impact of imperfect mask use on reproduction number of COVID-19: a next generation matrix approach. Infect Dis Model. 2020;5:405–408. doi: 10.1016/j.idm.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quilty BJ, Clifford S, Hellewell J, et al. Quarantine and testing strategies in contact tracing for SARS-CoV-2: a modelling study. Lancet Public Health. 2021;6:e175–e183. doi: 10.1016/S2468-2667(20)30308-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyre DW, Lumley SF, O'Donnell D, et al. Differential occupational risks to healthcare workers from SARS-CoV-2 observed during a prospective observational study. eLife. 2020;9 doi: 10.7554/eLife.60675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen I, Crozier A, Buchan I, Mina MJ, Bartlett JW. Recalibrating SARS-CoV-2 antigen rapid lateral flow test relative sensitivity from validation studies to absolute sensitivity for indicating individuals shedding transmissible virus. Clin Epidemiol. 2021;13:935–940. doi: 10.2147/CLEP.S311977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eyre DW, Taylor D, Purver M, et al. Effect of COVID-19 vaccination on transmission of alpha and delta variants. N Engl J Med. 2022;386:744–756. doi: 10.1056/NEJMoa2116597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee LYW, Rozmanowski S, Pang M, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity by viral load, S gene variants and demographic factors, and the utility of lateral flow devices to prevent transmission. Clin Infect Dis. 2022;74:407–415. doi: 10.1093/cid/ciab421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Institute of Population Health. University of Liverpool Covid-SMART asymptomatic testing pilot in Liverpool city region: quantitative evaluation. Dec 20, 2021. https://www.liverpool.ac.uk/media/livacuk/coronavirus/Liverpool_City_Region_Covid_SMART_Evaluation-Feb.pdf
- 18.Roix J, Sudhanva M, Cox T, et al. Evaluation of endpoint PCR (EPCR) as a central laboratory based diagnostic test technology for SARS-CoV-2. UK Government Department of Health and Social Care. Jan 28, 2021. https://www.gov.uk/government/publications/evaluation-of-endpoint-pcr-epcr-as-a-diagnostic-test-technology-for-sars-cov-2/evaluation-of-endpoint-pcr-epcr-as-a-central-laboratory-based-diagnostic-test-technology-for-sars-cov-2
- 19.UK Health Security Agency Investigation of SARS-CoV-2 variants: technical briefings. Feb 22, 2023. https://www.gov.uk/government/publications/investigation-of-sars-cov-2-variants-technical-briefings
- 20.Wasserstein RL, Lazar NA. The ASA statement on p-values: context, process, and purpose. Am Stat. 2016;70:129–133. [Google Scholar]
- 21.Schuit E, Venekamp RP, Hooft L, et al. Diagnostic accuracy of COVID-19 rapid antigen tests with unsupervised self-sampling in people with symptoms in the omicron period: cross sectional study. BMJ. 2022;378 doi: 10.1136/bmj-2022-071215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osterman A, Badell I, Basara E, et al. Impaired detection of omicron by SARS-CoV-2 rapid antigen tests. Med Microbiol Immunol (Berl) 2022;211:105–117. doi: 10.1007/s00430-022-00730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soni A, Herbert C, Filippaios A, et al. Comparison of rapid antigen tests' performance between delta and omicron variants of SARS-CoV-2: a secondary analysis from a serial home self-testing study. Ann Intern Med. 2022;175:1685–1692. doi: 10.7326/M22-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landaas ET, Storm ML, Tollånes MC, et al. Diagnostic performance of a SARS-CoV-2 rapid antigen test in a large, Norwegian cohort. J Clin Virol. 2021;137 doi: 10.1016/j.jcv.2021.104789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parvu V, Gary DS, Mann J, et al. Factors that influence the reported sensitivity of rapid antigen testing for SARS-CoV-2. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.714242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jääskeläinen AE, Ahava MJ, Jokela P, et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. J Clin Virol. 2021;137 doi: 10.1016/j.jcv.2021.104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapari A, Braliou GG, Papaefthimiou M, et al. Performance of antigen detection tests for SARS-CoV-2: a systematic review and meta-analysis. Diagnostics (Basel) 2022;12 doi: 10.3390/diagnostics12061388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deeks JJ, Singanayagam A, Houston H, et al. SARS-CoV-2 antigen lateral flow tests for detecting infectious people: linked data analysis. BMJ. 2022;376 doi: 10.1136/bmj-2021-066871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García-Fiñana M, Hughes DM, Cheyne CP, et al. Performance of the Innova SARS-CoV-2 antigen rapid lateral flow test in the Liverpool asymptomatic testing pilot: population based cohort study. BMJ. 2021;374 doi: 10.1136/bmj.n1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UK Health Security Agency LFD digital reader. Evaluation of real world deployment: final report. July, 2022. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1115461/LFD-digital-reader-november-2022.pdf
- 31.Consortium TLA, Beggs AD, Caiado CCS, et al. Machine learning for determining lateral flow device results for testing of SARS-CoV-2 infection in asymptomatic populations. Cell Rep Med. 2022;3 doi: 10.1016/j.xcrm.2022.100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data collected for the study, including de-identified individual participant data and a data dictionary defining each field in the set, will be available from NHS Digital's Data Access Request Service with publication of this Article. Details of how to apply for access to the data via the Data Access Request Service are provided at https://digital.nhs.uk/services/data-access-request-service-dars.