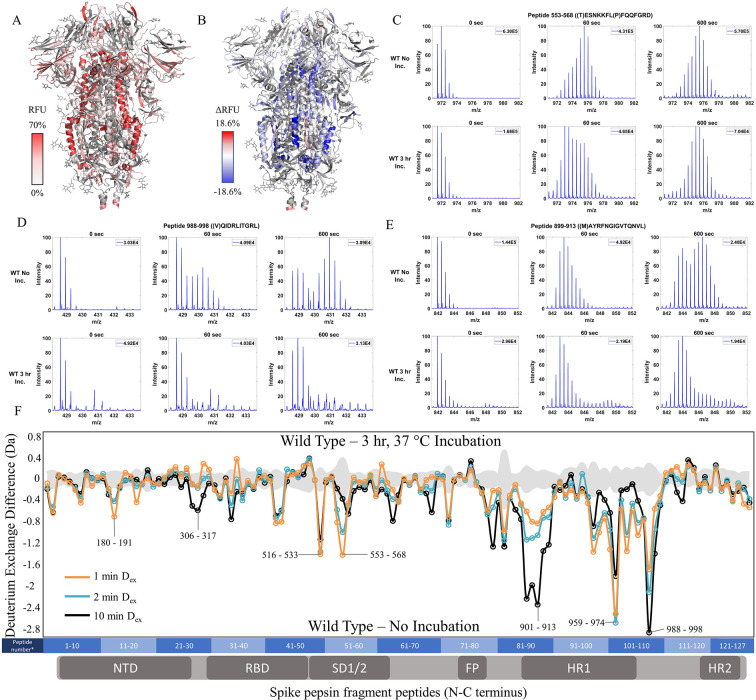
Figure 2. Prefusion conformation for wild-type (WT) spike (S) protein favored by 3 hr incubation at 37℃.
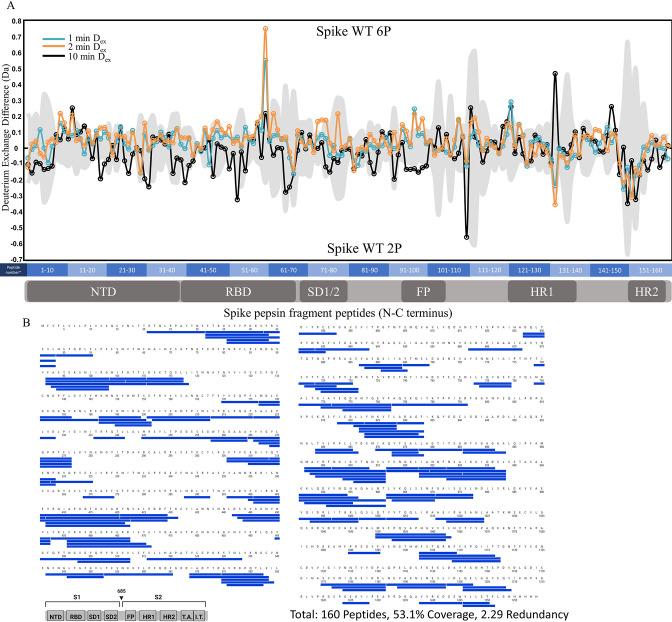
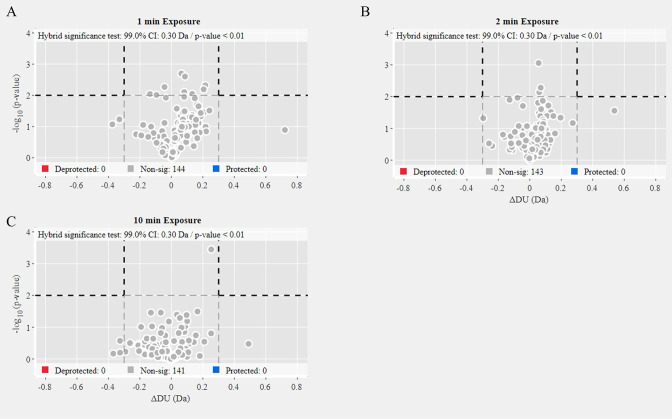
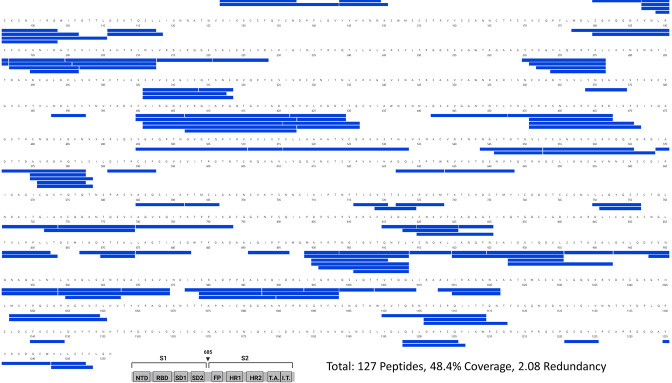
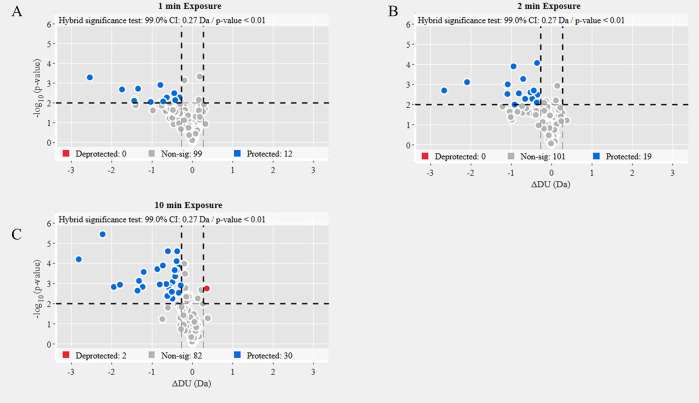
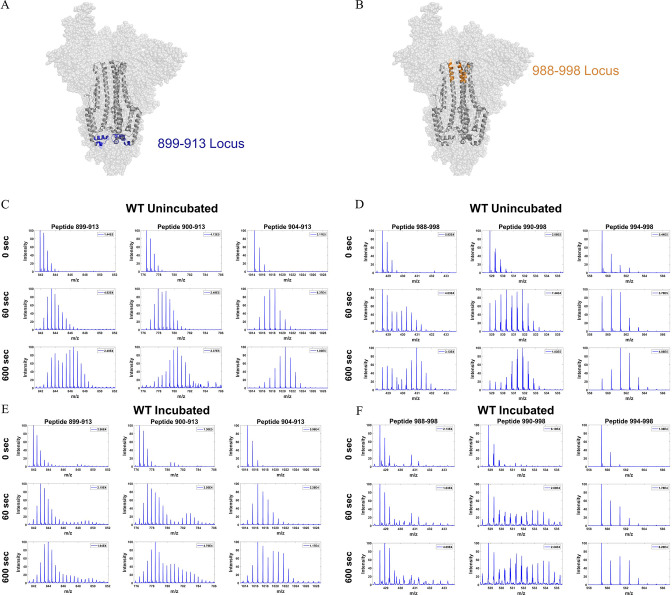
(A) Relative fractional uptake (Dex = 10 min) for unincubated WT S mapped onto an S trimer structure with three ‘down’ RBDs (PDB ID: 6VXX) (coverage of WT S constructs shown in Figure 2—figure supplements 1–2, differences in deuterium exchange for WT 2P and 6P constructs shown in Figure 2—figure supplement 3). Deuterium exchange heat map gradient of white (0%) -red (70%) as mapped on S structure (PDB ID: 6VXX). (B) Differences in deuterium exchange (Δ RFU) (Dex = 10 min) for WT S after a 3 hr incubation at 37℃ minus unincubated WT S were mapped onto the S structure (PDB ID: 6VXX). Shades of blue correspond to negative differences in deuterium exchange and shades of red correspond to a positive difference in deuterium exchange. (C–E) Stacked mass spectra for WT S peptides 553–568, 899–913, and 988–998 with undeuterated reference spectra, 1 min and 10 min exchange (left to right). For each peptide, the top row shows spectra for unincubated WT S and the bottom row shows spectra for WT S incubated for 3 hr at 37°C. Absolute intensities are indicated at the top right of each spectrum. (F) Differences in deuterium exchange (deuterons) mapped at peptide resolution from N to C terminus for WT S incubated for 3 hr at 37°C minus unincubated WT S are shown in difference plots for 1-, 2-, and 10 min exchange. Select peptides showing significant differences in exchange are annotated. Significance was determined by hybrid significance testing (p<0.01, Figure 2—figure supplement 4). Differences are tabulated in Figure 2—source data 1 with corresponding peptide numbers* shown on the x-axis of the difference plot.