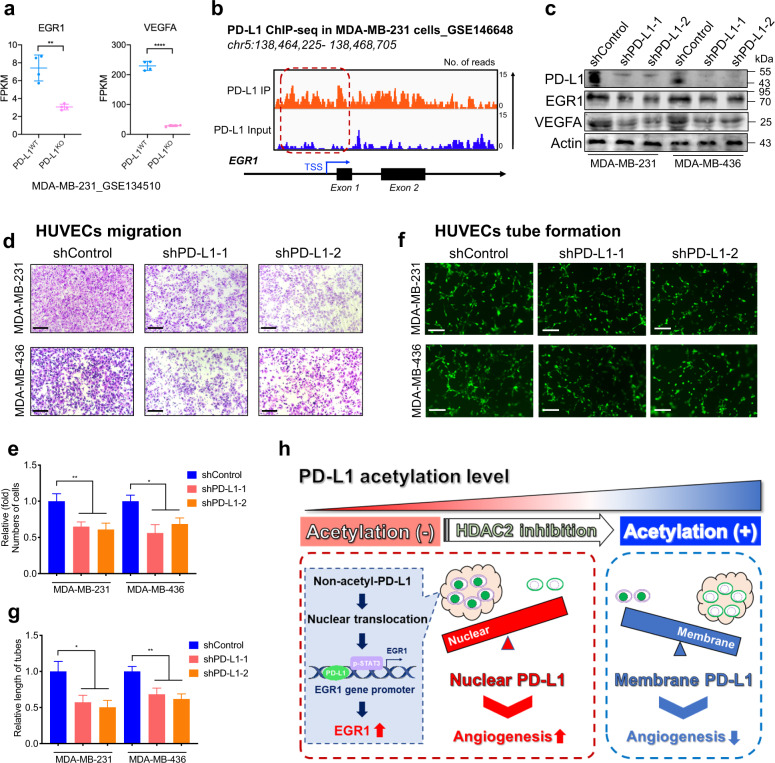
Fig. 8. Nuclear PD-L1 enhanced EGR1 expression and angiogenesis in breast cancer.
a FPKM levels of EGR1 and VEGFA from RNA-seq analysis in breast cancer cell line MDA-MB-231 after PD-L1 knockout (GSE134510). n = 4. Data are presented as means ± SD. Two-tailed unpaired Student’s t-test. b IGV tracks for EGR1 from PD-L1 ChIP-seq analysis in breast cancer cell line MDA-MB-231 (GSE146648). c Western blot images showing the protein levels of PD-L1, EGR1 and VEGFA in PD-L1-silenced breast cancer cells and control cells. d HUVECs migration assay to investigate the effect of conditional medium from PD-L1-silenced breast cancer cells and control cells on the migration ability of HUVECs. Scale bars, 50 µm. e Statistical analysis of the HUVECs migration assay. n = 3. Data are presented as means ± SD. Two-tailed unpaired Student’s t-test. f HUVECs tube formation assay to investigate the effect of conditional medium from PD-L1 silenced breast cancer cells and control cells on HUVECs tubule formation. Scale bars, 50 µm. g Statistical analysis of the HUVECs tube formation assay. n = 3. Data are presented as means ± SD. Two-tailed unpaired Student’s t-test. h Schematic diagram of pro-angiogenic mechanism of nPD-L1 in UM. We demonstrated that PD-L1 nuclear translocation in UM facilitated angiogenesis by coactivating EGR1 expression with p-STAT3. This elevated nPD-L1 level was mediated by HDAC2-dependent deacetylation in UM cells.