Figure 6. Nr5a2 regulates jaw-specific enhancer accessibility and gene expression for perichondrium and tendon genes.
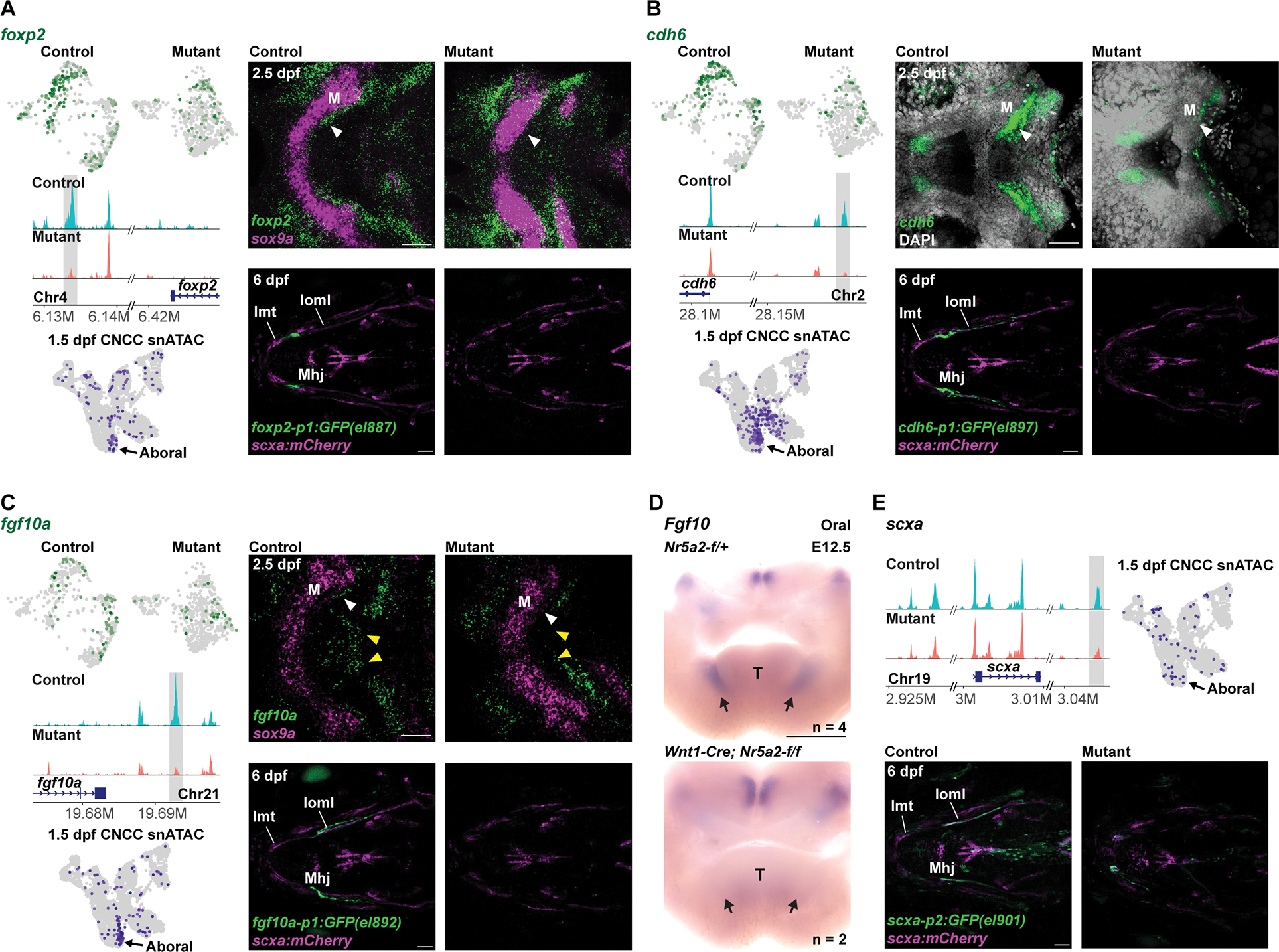
(A-C) For each gene, feature plots in top left show downregulation of transcripts in mutant (nr5a2:mGFP-DBD-del/oz3) versus control (nr5a2:mGFP-DBD-del/+) mandibular mesenchyme. Middle left shows genomic tracks of chromatin accessibility, with the Nr5a2 motif-harboring regions with decreased accessibility shown in grey. Bottom left shows feature plots of 1.5 dpf CNCC snATAC data 17 highlighting accessibility of the shaded regions in the aboral mandibular domain. Top right shows fluorescent in situ hybridizations of the mandibular and hyoid arches in ventral views, with sox9a expression labeling chondrocytes (A, C) or DAPI labeling all nuclei (B). White arrowheads denote loss of perichondrium expression in mutants, and yellow arrowheads loss of midline fgf10a expression. Bottom right shows transgenic lines in which GFP is driven by the highlighted genomic regions. For each line, GFP expression is observed posterior to Meckel’s (M) cartilage and partially overlapping with scxa:mCherry in 5/5 wild types and completely lost in 5/5 mutants. Intermandibular tendon (Imt), interopercular–mandibular ligament (Ioml), mandibulohyoid junction tendon (Mhj).
(D) Oral view of in situ hybridizations for Fgf10 in dissected mandibular arches from control (Nr5a2-f/+) and mutant (Wnt1-Cre; Nr5a2-f/f) mouse embryos at E12.5. Fgf10 expression is selectively lost in mandibular domains (arrows). T, tongue.
(E) Top left shows genome tracks of chromatin accessibility at the scxa locus, with the region containing Nr5a2 motifs and having decreased accessibility in mutants highlighted in grey. At top right, feature plot from 1.5 dpf CNCC snATAC data shows accessibility of this region in the aboral mandibular domain. At bottom, transgenic line in which GFP is driven by the highlighted genomic region shows expression in the scxa:mCherry+ Ioml ligament in 5/5 wild types and loss of Ioml expression in 5/5 nr5a2oz3/oz3 mutants. Scale bars = 50 μm (A-C, E); 500 μm (D). See also Figure S6 for additional examples of transgenic line expression and expanded genomic views of open chromatin.
