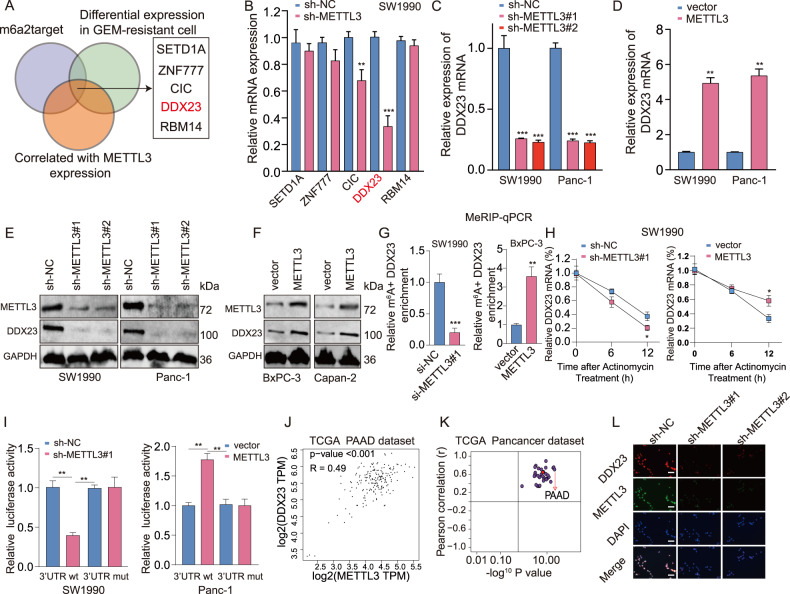
Fig. 3. The identification and validation of a direct downstream target of METTL3 in pancreatic cancer under gemcitabine resistance.
A The intersection between the list of predicted m6A targets of METTL3 by m6A2target, the list of differentially expressed genes in GEM-resistant pancreatic cancer, and the list that contained genes correlated with METTL3 expression showed the potential direct downstream targets of METTL3 in pancreatic cancer. B The relative mRNA expression of the identified five genes: SETD1A, ZNF777, CIC, DDX23, and RBM14 in cells with METTL3 silence. **P < 0.01, ***P < 0.001, compared with the sh-NC group. C, D The expression of DDX23 mRNA in the two cell lines with METTL3 knockdown (C) and METTL3 overexpression (D) was analyzed by qRT-PCR. C ***P < 0.001, compared with the sh-NC group. D **P < 0.01, compared with the vector group. E, F The expression of DDX23 protein in the two cell lines with METTL3 knockdown (E) and METTL3 overexpression (F) was determined using western blotting. G The m6A level of DDX23 in pancreatic cell lines with METTL3 knockdown (left panel) and overexpression (right panel) was measured. ***P < 0.001, compared with the sh-NC group; **P < 0.01, compared with the vector group. H The half-life of DDX23 mRNA in cells with respective METTL3 knockdown and overexpression with Actinomycin treatment was analyzed using qRT-PCR. I The regulatory relationship between METTL3 and DDX23 mRNA was validated using the dual-luciferase reporter gene assay. J In TCGA pancreatic cancer data, METTL3 and DDX23 were found significantly positively correlated. PAAD: pancreatic adenocarcinoma. K In all the cancers of TCGA, METTL3 expression was found significantly correlated with DDX23 expression. L The FISH assay showed that METTL3 knockdown led to DDX23 downregulation in the pancreatic cancer cell line.