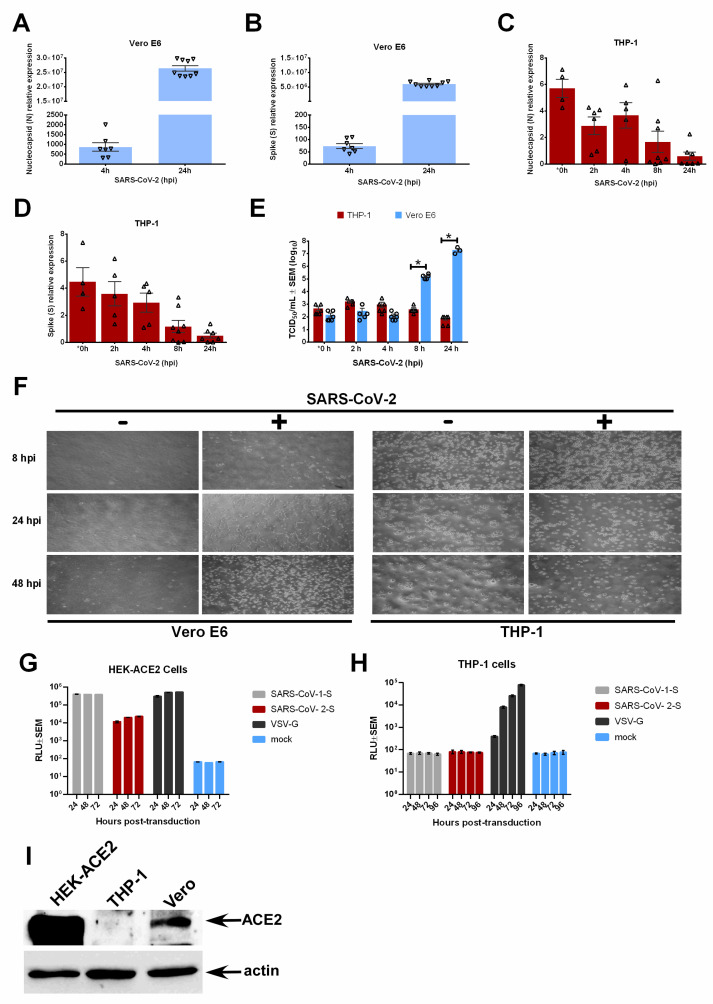
Figure 2.
RT-qPCR was used to detect SARS-CoV-2 nucleocapsid or N (A) and Spike or S (B) viral genes in SARS-CoV-2-infected Vero E6 cells. RT-qPCR was used to detect N (C) and S (D) viral genes in SARS-CoV-2-infected human THP-1 macrophages. (E) Culture supernatants from SARS-CoV-2-infected human THP-1 macrophages and Vero E6 cells were analyzed by TCID50 assay to determine infectious virus production. (F) Bright field microscopy photographs of Vero E6 (MOI = 0.1) and THP-1 (MOI = 0.5) macrophages infected with SARS-CoV-2 for the indicated times. Luminescence (RLU) was measured to determine transduction of HEK-ACE2 cells (HEK293T cells stably expressing human ACE2) (G) and THP-1 macrophages (H) by luciferase-expressing lentiviruses pseudotyped with VSV glycoprotein (VSV-G), SARS-CoV-1 S protein, or SARS-CoV-2 S protein. (I) ACE2 expression was analyzed in HEK-ACE2, THP-1, and Vero cells by Western blotting with ACE2 and actin (loading control) antibodies. Error bars denote the standard error of the mean (SEM) from 2 to 3 biologically independent experiments. hpi = hours post-infection. * p < 0.05 was determined by two-way ANOVA adjusted by Sidak’s multiple comparison test. * 0 h timepoint data were collected 1 h post-virus adsorption.