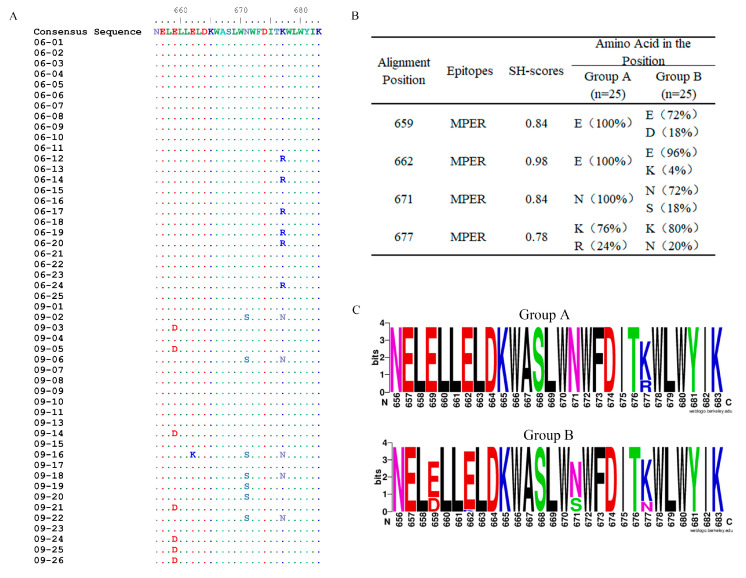
Figure 2.
Comparisons of the variation of Env between groups A and B and amino acid sequences with names consistent with nucleic acid sequences. (A) Amino acid mutations in the MPER (656–683) at two time points with positions corresponding to HXB2. • (point) represents the same residues with consensus sequence. (B) SH values of the mutated positions in the MPER. (C) Sequence logos of aa signatures.