Fig. 1.
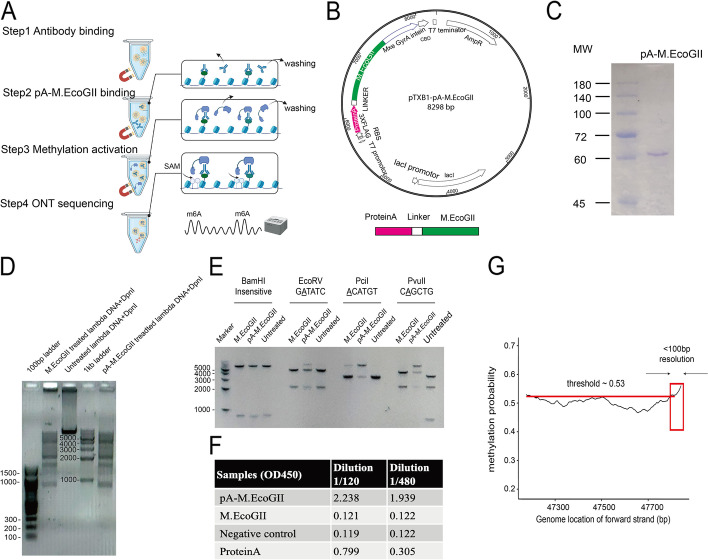
The experiment concept and in vitro validation of the recombinant protein pA-M.EcoGII in BIND&MODIFY. A The experiment outlines of BIND&MODIFY. After light fixation and permeabilization, the cells were tethered to Concanavalin A magnetic beads (which bind specifically to mannosyl- and glucosyl-containing extracellular glycoproteins on the cell membrane) for purification in the following steps. Step1 Antibody binding; Step 2 pA-M.EcoGII binding; Step 3 Methylation activation; Step 4 ONT sequencing. Firstly, recombinant protein pA-M.EcoGII was tethered to desired sites under antibody guidance. Then the M.EcoGII m6A methylation activity was locally activated with the addition of S-adenosylmethionine to modify the nearby regions to label the genomic DNA with targeted binding proteins. Finally genomic DNA was extracted to prepare the library for ONT nanopore sequencing. After sequencing, the data was processed as genome alignment and m6A base calling (refer to detail protocol). B The upper panel showed the plasmid map of the pA-M.EcoGII. The fused pA-M.EcoGII was cloned into pTXB1 plasmid and purified with compatible IMPACT protein purification system. The lower panel showed the expressed fusion protein structure: Protein A-linker-M.EcoGII-intein-CBD. C The Coomassie blue gel stain showed the purity of the purified pA-M.EcoGII. D Methylation of linear lambda DNA by pA-M.EcoGII activates m6A-site-dependent DpnI restriction endonuclease digestion. The PCR amplified unmethylated lambda DNA was treated with commercial M.EcoGII, no enzyme, and pA-M.EcoGII. The GATC m6A methylation-dependent restriction endonuclease DpnI digestion suggested the comparable methyltransferase activity of the commercial M.EcoGII and our recombinant proteins. DNA marker: 100 bp ladder, 100–1510 bp(left); 1 kb ladder, 250–10,000 bp(right). E Methylation of linear dsDNA by pA-M.EcoGII inhibits multiple site-specific methylation-sensitive restriction endonucleases. The unmethylated DNA template was a 7-kb linear dsDNA, which was PCR amplified from pTXB1 plasmid. The DNA template was treated with commercial M.EcoGII, pA-M.EcoGII, and no enzyme. These treated DNA templates were each incubated with four restriction endonucleases (BamHI, EcoRV, PciI, PvuII). The BamHI is the m6A methylation insensitive enzyme, and the EcoRV, PciI, and PvuII are the m6A methylation-sensitive enzyme, with which the digestion could be blocked by corresponding m6A site. Our pA-M.EcoGII recombinant protein showed digestion inhibition on EcoRV, PciI, and PvuII digested samples, better than commercial M.EcoGII, as compared to untreated DNA template. DNA marker, 1 kb ladder, 250–10,000 bp. F The antibody affinity assay showed the recombinant pA-M.EcoGII had the affinity to the secondary antibody in two different dilutions (1/120, 1/480, 10 mg/ml). G In vitro validation of BIND&MODIFY resolution with single base 5mC containing lambda DNA. A fragment of 700 bp lambda DNA was amplified by PCR, and 5mC was introduced near the end of forward strand only by the modified primers with the precise 5mC site. The 5mC labeled DNA was bound by 5mC antibody and was subsequently treated by BIND&MODIFY method. The m6A probability (Megalodon calling probability) of forward strand was calculated and plotted with loci. When the methylation probability cut-off was set at 0.53, the high methylation probability region was observed in 3′ end, overlapping with the expected the 5mC site