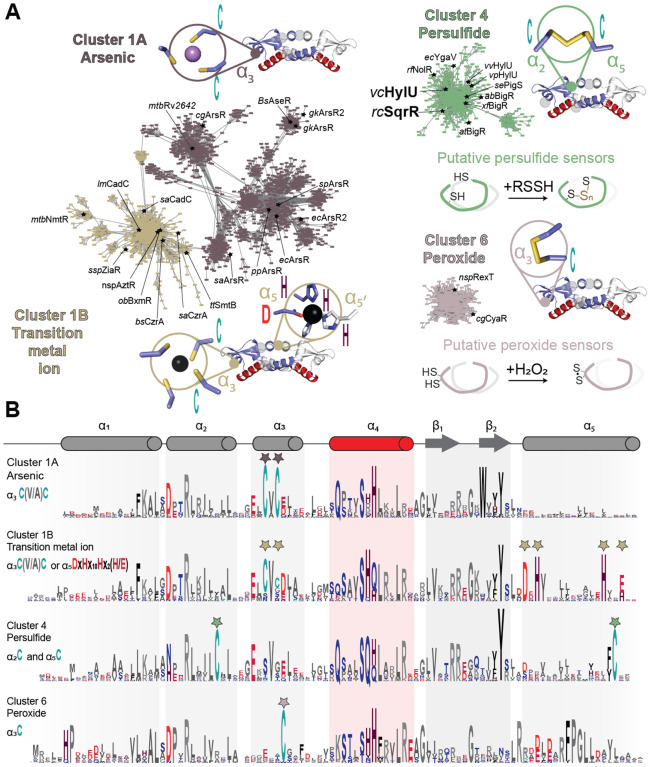
Figure 1.
Sequence similarity network (SSN) analysis of the ArsR superfamily of bacterial repressors. (A) Results of an SSN clustering analysis of 168,163 unique sequences belonging to the Pfam PF01022 and Interpro IPR001845 Pfam using genomic enzymology tools and visualized using Cytoscape. Clusters functionally annotated as arsenic (As), nickel/cobalt (Ni/Co), persulfide (RSSH) or hydrogen peroxide (H2O2) sensors are presented here, and all the determined main clusters are presented in Fig. S1. All clusters are designated by a number and ranked according to the number of unique sequences (Table S2), color-coded, and, if known. Each node corresponds to sequences that are 50% identical over 80% of the sequence, using an alignment score of 22 (see Materials and Methods). Functionally characterized members in each are indicated with species name and trivial name. (B) Sequence logo representations of sequence conservation defined by the indicated cluster of sequences derived from panel A. The residues that coordinate metals/metalloid ions in ArsR are marked by stars and the residues that undergo redox chemistry are marked with arrows. WebLogos other clusters are also provided (Figure S1B).