Figure 3:
Modeling of lipid-driven mechanical changes in the IMM and their homeostatic responses.
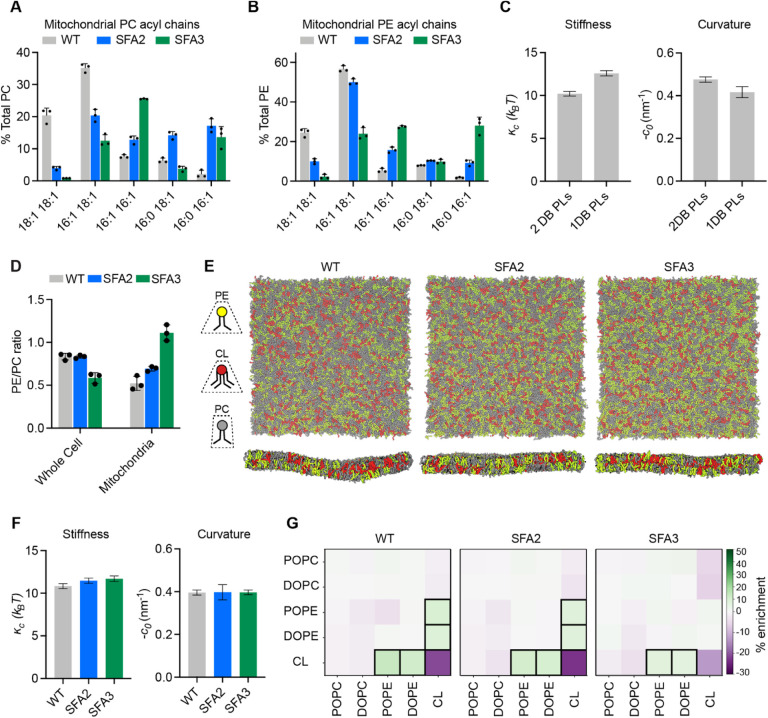
(A) Lipidomic profile of isolated mitochondria showing a transition from di-unsaturated to monounsaturated PC, n=3 biological replicates. Error bars represent SD.
(B) Mitochondrial PE shows a similar transition from di-unsaturated species to monounsaturated species, n=3 biological replicates. Error bars represent SD.
(C) Changes to membrane mechanical properties predicted by Martini 2 CG-MD simulations of mitochondrial-like lipid mixtures that shift from di-unsaturated (2 double bonds, DB) to mono-unsaturated (1 DB) PLs. Stiffness (bending modulus) increases while negative spontaneous curvature, derived from the first moment of the lateral pressure profile and bending modulus, decreases. Compositions of these ‘ideal’ systems are shown in Figure EV3A.
(D) Increasing lipid saturation reduces the PE/PC ratio in whole cells, but increases it in isolated mitochondria, suggesting a curvature-based adaptation to increasing saturation.
(E) Top-down and side-on snapshots of CG-MD bilayers showing headgroup adaptation to increasing saturation in SFA strains.
(F) Simulations of the lipid systems derived from SFA mitochondria that incorporate homeostatic head-group changes. In contrast to the systems in (C), the complex systems show only small changes to membrane mechanical properties, suggesting that headgroup adaptations may offset the changes to mechanical properties associated with increased saturation.
(G) Compositional enrichment within the local neighborhood of individual lipid types. Given a lipid type (y-axis) the color indicates the percent enrichment in likelihood of observing an x-axis labeled lipid within a 1.5 nm radius neighborhood compared to random distribution. Boxes indicate conditions with greater than 5% enrichment from random. PO/DOPE are enriched around CL, indicating the association between conically shaped lipids.