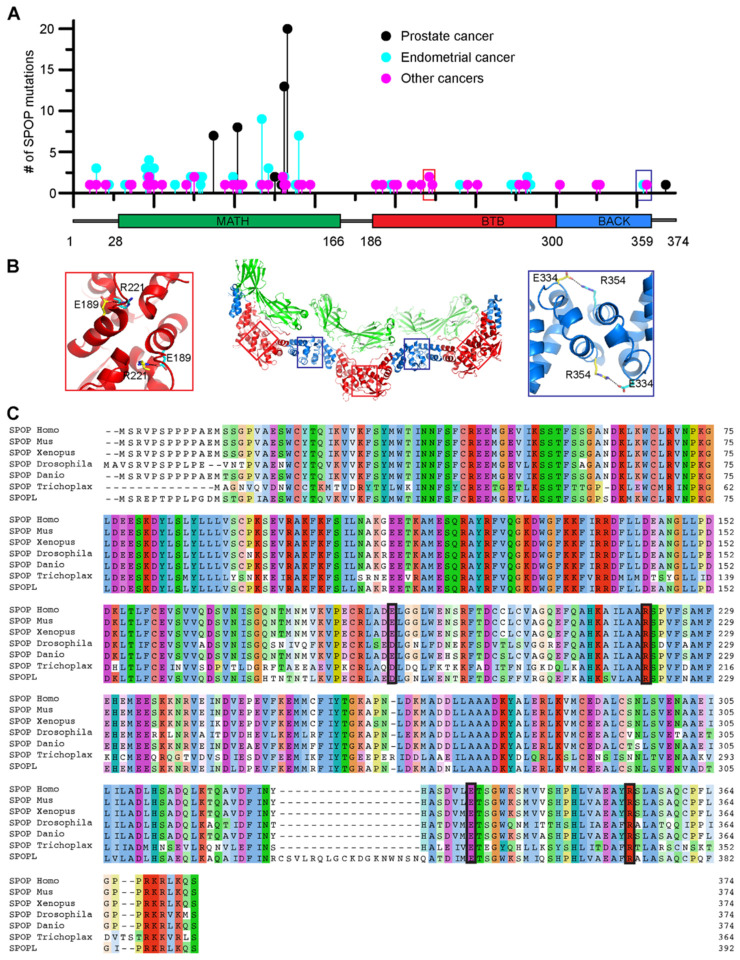
Figure 2. Conserved residues that form contacts across the dimerization interfaces are mutated in cancer patients.
(A) The lollipop plot shows mutations identified in cancer patients in all three domains of SPOP. Prostate cancer–associated mutations (black) are prevalent in the substrate-binding cleft of the MATH domain. These mutations impair SPOP–substrate interactions, resulting in the stabilization of oncoproteins in prostate cancer patients. Endometrial cancer mutations (blue), which have intermediate frequency, are also clustered in the MATH domain. However, these surprisingly promote ubiquitination of some substrates while inhibiting ubiquitination of others. Among the other SPOP mutations identified (magenta), R221C (red box) and R354H (blue box) are located in the BTB and BACK domains, respectively. Mutation data were collated from cBioPortal (33, 34). (B) Model of a SPOP oligomer generated by superimposing the SPOP crystal structures for the BTB dimer (35) and BACK dimer (36). Red and blue boxes show the locations of R221 and R354 in the BTB/BTB and BACK/BACK interfaces, respectively. Both residues form salt bridges with glutamic acid residues across their respective dimerization interfaces. (C) Sequence alignment of human SPOP with SPOP homologs in other species and with human SPOPlike protein (SPOPL). The alignment shows evolutionary conservation of R221 and R354 and their corresponding salt bridge partners E189 and E334. Homo, Homo sapiens; Mus, Mus musculus; Xenopus, Xenopus laevis; Drosophila, Drosophila melanogaster; Danio, Danio rerio; and Trichoplax, Trichoplax adhaeren.