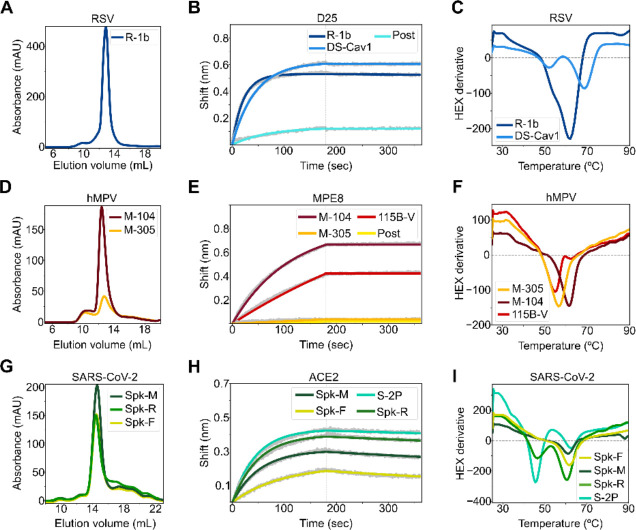
Figure 2. Biochemical characterization of designed variants.
(A) Size-exclusion chromatography (SEC) of monodispersed RSV F designs. (B) Binding of design R-1b to the prefusion-specific antibody D25 compared to the clinical candidate DS-Cav1 and the postfusion RSV A2 F (post). (C) Differential scanning fluorimetry (DSF) of design R-1b and the clinical candidate DS-Cav1. DS-Cav1 was used to compare the stability of R-1b as the parental sequence of the latter is not prefusion-stabilized. (D) SEC of monodispersed hMPV F designs. (E) Binding of designed hMPV F variants to the prefusion-specific antibody MPE8 compared to their parent prefusion construct 115B-V and the postfusion hMPV B2 F (post). (F) DSF of designed hMPV F variants and their parent prefusion construct 115B-V. (G) SEC of monodispersed SARS-CoV-2 S designs. (H) Binding of designed SARS-CoV-2 S variants to ACE2 compared to their parent prefusion construct S-2P. (I) DSF of designed SARS-CoV-2 S variants and their parent prefusion construct S-2P. Antibody binding assays show in grey the raw data, in colors the fitted curves, and in dotted lines the end of the association time. Binding constants are shown in Tables S2, S3, and S4.