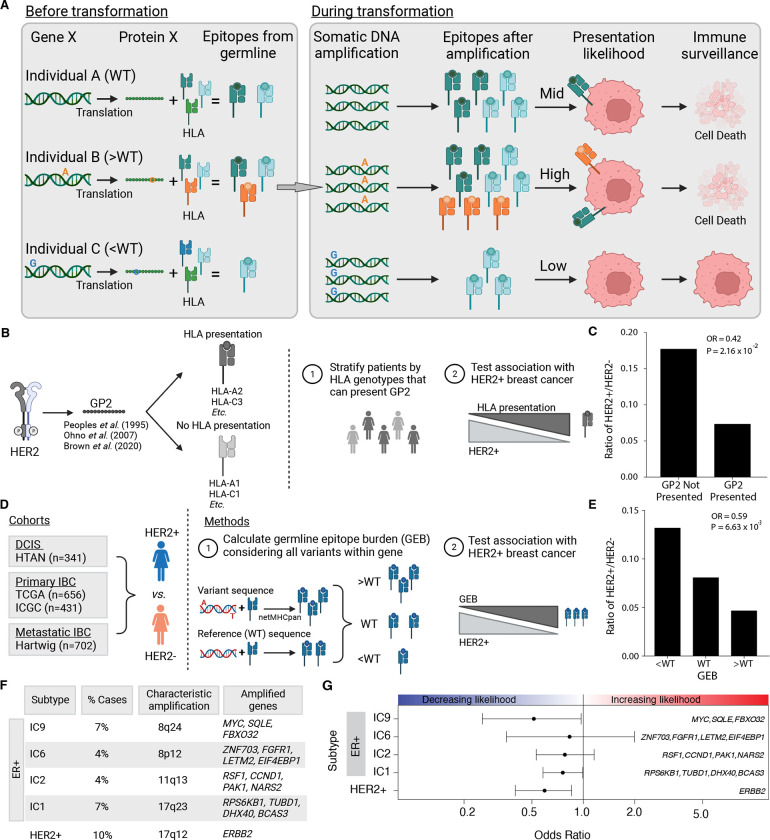
Figure 1 – Germline-derived epitope burden in oncogenes selects against oncogene amplification.
A) Schematic of germline-mediated immunoediting. Prior to transformation, differing germline genomes and HLA alleles result in differing numbers of epitopes derived from a gene of interest (e.g. Gene X). If during transformation, the tumor acquires additional copies of Gene X (i.e. somatic amplification), the number of epitopes increases further. As a result, individuals with a high burden of epitopes are more likely to be surveilled by the immune system triggering cell death. B) As proof of concept, GP2 is a well-characterized, naturally occurring (i.e., non-somatically mutated) immunogenic peptide derived from HER2. Schematic overview of analysis framework to investigate if the ability to present GP2, i.e., having MHC Class I alleles that bind GP2, is associated with HER2+ breast cancer. C) The ability to present GP2 is negatively associated with HER2+ breast cancer. Barplot shows the ratio of HER+ to HER2− in patients that have HLA alleles that can bind GP2 (GP2 presented) vs patients that do not (GP2 not presented). Odds ratio (OR) and p-value from logistic regression model correcting for first six genetic principal components. D) Schematic outlining methods to investigate germline-mediated immunoediting. Using four independent cohorts representing multiple stages of breast cancer, pre-invasive, primary invasive and metastatic invasive breast cancer, we investigated if the GEB in a gene of interest was associated with the likelihood of acquiring a somatic amplification of the gene using HER2 as a representative example. E) GEB in ERBB2 is negatively associated with HER2+ breast cancer. Barplot shows the ratio of HER2+ to HER2− patients with low, medium or high GEB. Odds ratio (OR) and p-value from logistic regression model correcting for the first six genetic principal components. F) Beyond ERRB2, we investigated amplicons that characterize four ER+/HER2− high risk of relapse subtypes (IC1, IC2, IC6 and IC9), where the percent of breast cancer cases they represent and the corresponding chromosome region and core genes is denoted for each subtype. G) GEB in recurrently amplified genes is negatively associated with gene amplification. Scatterplot shows odds ratio (x-axis) and 95% confidence intervals from logistic regression model correcting for the first six genetic principal components and somatic mutation burden. Covariates in the top panel indicate the direction of the effect, namely whether GEB is associated with increased or decreased likelihood of each subtype.