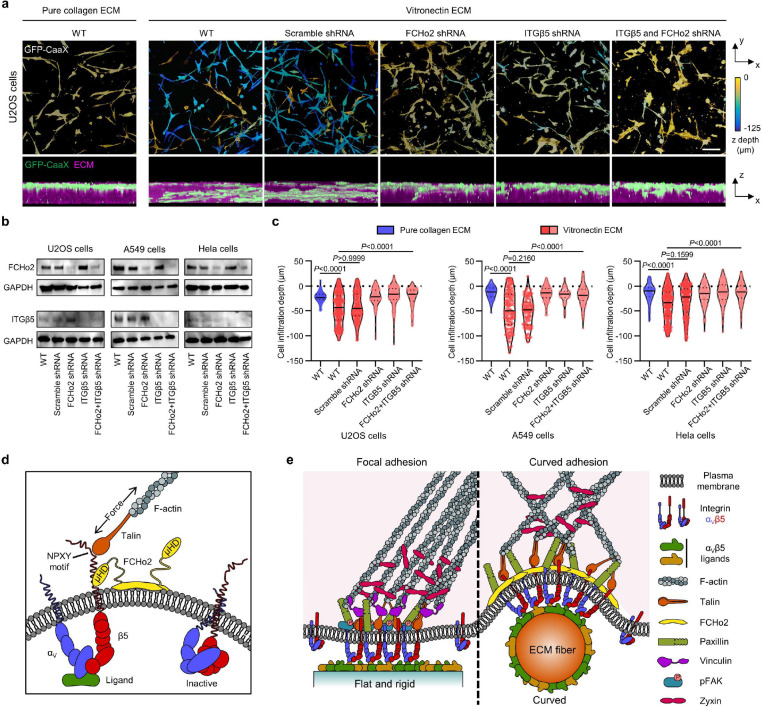
Fig. 6: Curved adhesions facilitate the cell migration in 3D ECMs.
a, Representative images of U2OS cells expressing GFP-CaaX in 3D matrices after 72-hr culture. Cells are colour-coded according to their depth in the matrices in x-y projections. Cells are coloured in green and merged with ECM in magenta in x-z projections. WT cells infiltrated into matrices made of vitronectin fibres, but not matrices made of pure collagen fibres. The shRNAs of FCHo2, ITGβ5, or both, but not scramble shRNA inhibited the cell infiltrations. b, Western blots show that shRNAs are able to effectively reduce the expression of FCHo2, ITGβ5, or both in U2OS, A549, and Hela cells. These results support the knock-down efficiencies supplied by the company (Supplementary Table 1). c, Quantification of the cell infiltration depth for U2OS, A549, and Hela cells in 3D pure collagen matrices or 3D vitronectin matrices with the knockdown of FCHo2, ITGβ5, or both. N = 100–300 cells for each condition, from two independent experiments. Medians (lines) and quartiles (dotted lines) are shown. P values calculated using Kruskal-Wallis test with Dunn’s multiple-comparison. d, Illustration of ITGβ5 interacting with FCHo2 in curved adhesions. e, Schematic comparison of focal adhesion and curved adhesion architectures. Scale bars: 100 μm.