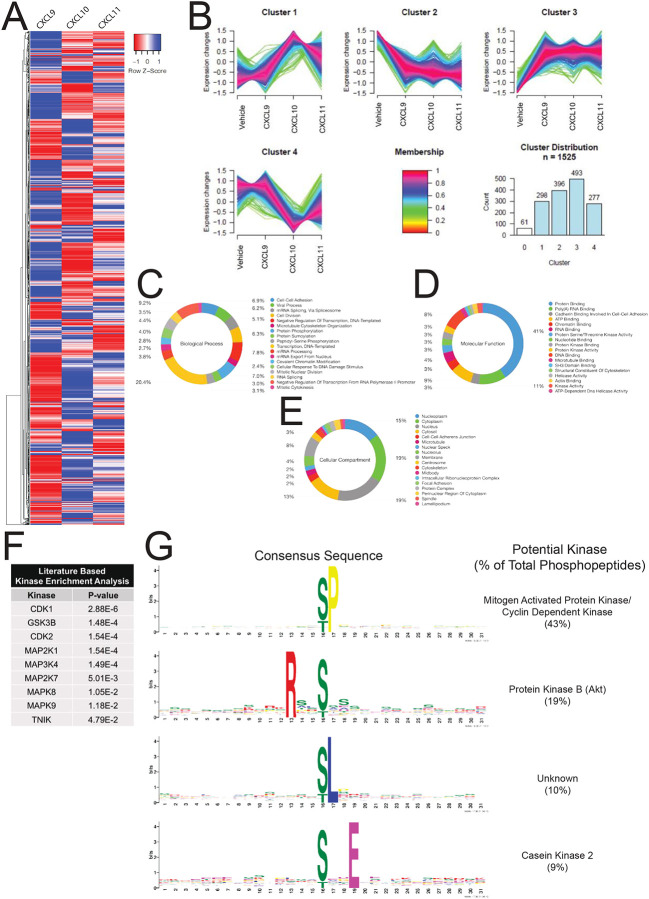
Figure 5: Characterization of the global phosphoproteome in HEK293 cells treated with endogenous CXCR3 agonist.
(A) HEK293 cells expressing CXCR3-WT were stimulated with vehicle control or 100 nM chemokine for five minutes. Heat map of statistically significant phosphopeptides normalized to vehicle control are shown. n=2 technical replicates of six pooled biological replicates. (B) Cluster analysis of significant phosphopeptides using GProX (Rigbolt et al., 2011). Cluster 0 is not shown for clarity due to low membership count. (C-E) Gene Ontology analysis of significant phosphopeptides as grouped by biological process, molecular function or cellular compartment, respectively. Percentiles demonstrate number of individual phosphopeptides present in each Gene Ontology Term. (F) Manually curated, literature-based kinase enrichment analysis to predict kinase activity based on significant phosphopeptides using Kinase Enrichment Analysis 2 (Lachmann and Ma’ayan, 2009). (G) Consensus sequences of significant phosphopeptides in the dataset as generated using MoMo from MeMe suite and identified kinases with listed consensus motif based on manual literature review (Keshava Prasad et al., 2009). See S7 for additional global phosphoproteomic data.