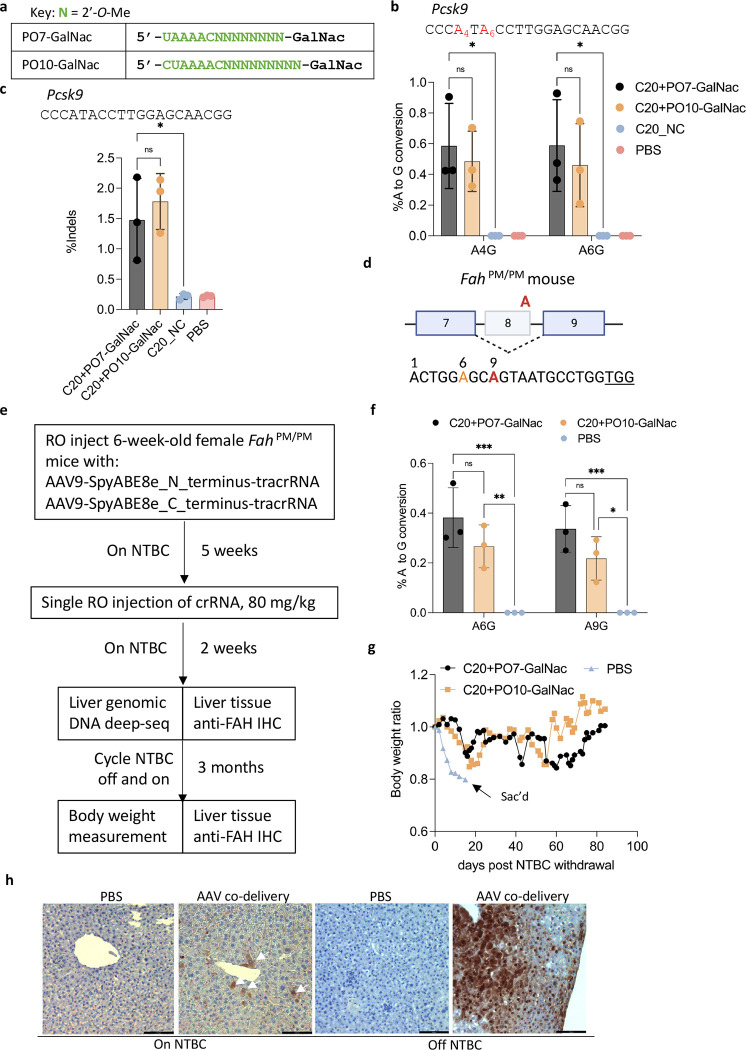
Figure 5. In vivo genome editing in the mouse liver following systemic administration of AAV vectors expressing effector + tracrRNA, co-delivered with C20 and GalNac-conjugated P.O.s.
a). Chemical modifications of the P.O.s used in this experiment. GalNac, trivalent N-acetylgalactosamine. b). Targeted amplicon deep sequencing quantifying the editing efficiency in the liver by co-delivery of AAV-expressed SpyCas9-ABE8e and tracrRNA, and a single dose of C20 with GalNac-conjugated P.O. at 80 mg/kg. C20_NC, C20 with a non-targeting spacer sequence (n = 3 mice per group). Data represent mean ± SD; ns, P > 0.05; *, P < 0.05, (two-way ANOVA). c). Targeted amplicon deep sequencing quantifying the editing efficiency in the liver by co-delivery of AAV-expressed SpyCas9 nuclease and tracrRNA, and a single dose of 80 mg/kg of C20 with GalNac-conjugated P.O. (n = 3 mice per group). Data present mean ± SD; ns, P > 0.05; *, P < 0.05, (one-way ANOVA). d). Schematic representation of the G-to-A point mutation that generates exon 8 skipping and Fah deficiency in the FahPM/PM mouse model. The bottom shows a previously validated guide design (44) for SpyCas9-ABE to correct the point mutation. Red, target adenine; orange, bystander adenine; underlined, PAM. e). Schematic representation of the experimental design of in vivo AAV co-delivery in the FahPM/PM mouse model. RO, retro-orbital injection; NTBC, 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3- cyclohexanedione. f). Base editing efficiency of the genomic DNA from mouse liver treated by AAV-SpyCas9-ABE-tracrRNA co-delivered with the indicated C20/P.O. compound. The editing efficiency was measured by targeted amplicon deep sequencing (n = 3 mice per group). Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-way ANOVA). g). Body weight of the FahPM/PM mice treated with AAV and the indicated crRNA, measured over 3 months of NTBC off-and-on cycles. h). IHC staining of mouse liver sections using an anti-FAH antibody. White arrows indicate FAH-positive hepatocytes. Scale bar, 100 μm.