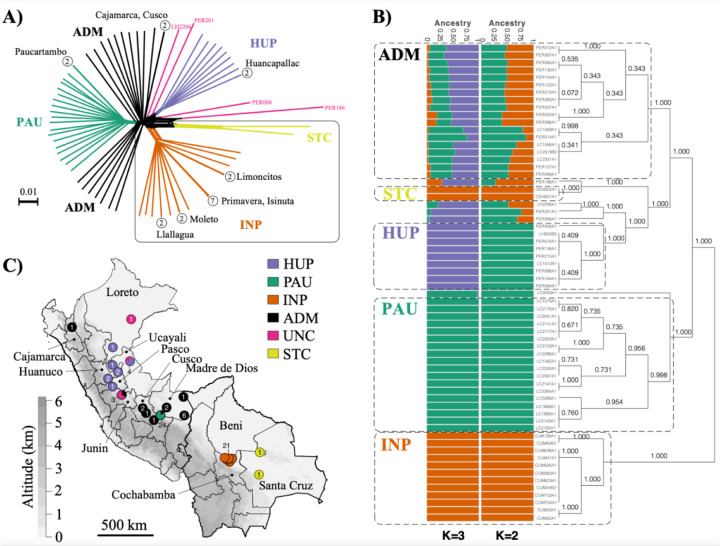
Figure 1. Population genomic structure and admixture in Lb parasites from Peru and Bolivia.
A) Phylogenetic network as inferred with SPLITSTREE based on uncorrected p-distances between 76 Lb genomes (excluding isolates CUM68, LC2318 and PER231) typed at 407,070 bi-allelic SNPs. Branches are coloured according to groups of Lb parasites as inferred with ADMIXTURE and fineSTRUCTURE (panel B). Box indicates the position of the Bolivian Lb genomes; all other genomes were sampled in Peru. Circles at the tips of seven branches point to groups of near-identical Lb genomes, showing the number of isolates and their geographical origin. B) ADMIXTURE barplot summarizing the ancestry components inferred at K = 2 or K = 3 populations in 65 Lb genomes. The phylogenetic tree summarizes the fineSTRUCTURE clustering results based on the haplotype co-ancestry matrix. Numbers indicate the MCMC posterior probability of a given clade. Dashed boxes indicate the three main parasite groups (INP, HUP, PAU) and the two main groups of admixed parasites (ADM, STC); the remainder of the parasites were of uncertain ancestry (UNC). C) Geographic map of Peru and Bolivia showing the origin of 76 Lb genomes. Dots are coloured according to groups of Lb parasites as inferred with ADMIXTURE and fineSTRUCTURE (panel B). Gray-scale represents altitude in kilometers, indicating the position of the Andes along the Peruvian and Bolivian Coast. Names are given for those Peruvian departments where a Lb parasite was isolated.