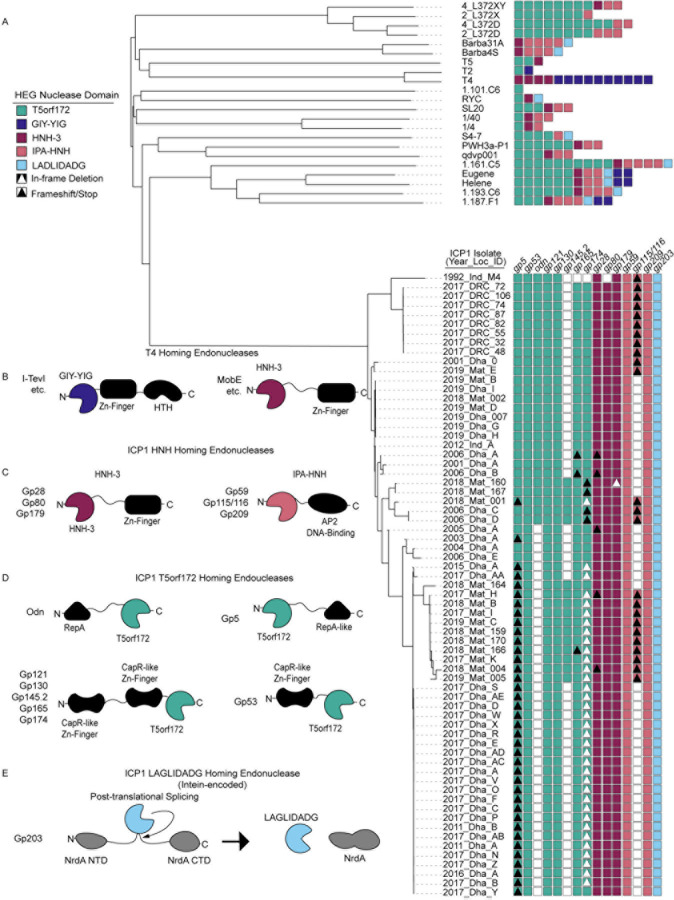
Figure 1. ICP1 phages encode many homing endonucleases and are enriched for T5orf172 genes.
(A) A phylogeny of the 67 ICP1 isolates (bottom clade) and various outgroup phages (top clade). ICP1 isolates are named according to the year of isolation, country of isolation, and an identifying character to differentiate phages co-isolated in the same year and location (e.g., 2017_Dha_A = 2017 isolate from Dhaka, Bangladesh, isolate A). A VipTree alignment of all genomes of interest and subsequent HMM profile predictions of homing endonuclease genes in each representative genome shows enrichment of T5orf172 homing endonucleases in the clade of ICP1 isolates compared to most outgroup phages. Notably, highly related ICP1 isolates encode variable homing endonuclease genes, with frequent frameshifting (black triangle), in-frame deletion (white triangle), and lack of HEG altogether (white box) represented. These predictions are likely underestimates of the full HEG repertoire among these phages due to the genetic drift of HEGs impeding functional predictions.
(B-E) The variable homing endonuclease domain architecture between T4-encoded (B) and ICP1-encoded homing endonucleases (C-E). The predicted DNA binding domains are shown as black shapes and the predicted nuclease domains are colored according to the legend in (A). Apart from the LAGLIDADG HEG gp203, which is intein encoded and post-translationally spliced out of the functional NrdA protein, each of the represented domain architectures shares the general pattern of one terminus encoding at least a single DNA-binding domain and the opposite terminus encoding the nuclease effector domain.