Abstract
Aging is accompanied by a host of social and biological changes that correlate with behavior, cognitive health and susceptibility to neurodegenerative disease. To understand trajectories of brain aging in a primate, we generated a multiregion bulk (N = 527 samples) and single-nucleus (N = 24 samples) brain transcriptional dataset encompassing 15 brain regions and both sexes in a unique population of free-ranging, behaviorally phenotyped rhesus macaques. We demonstrate that age-related changes in the level and variance of gene expression occur in genes associated with neural functions and neurological diseases, including Alzheimer’s disease. Further, we show that higher social status in females is associated with younger relative transcriptional ages, providing a link between the social environment and aging in the brain. Our findings lend insight into biological mechanisms underlying brain aging in a nonhuman primate model of human behavior, cognition and health.
Aging in animals is characterized by changes in physiology and behavior and is accompanied almost universally by declines in cognition, health and longevity. Additionally, the risk of developing neurodegenerative diseases, including Parkinson’s disease and Alzheimer’s disease (AD), increases with age, indicating that age-associated changes in the brain may underlie disease pathogenesis and that their identification may provide targets for designing earlier and more effective interventions1. Our knowledge of aging trajectories in the brain and their links to neurodegenerative diseases and environmental risk factors is currently limited to studies in laboratory animal models with substantially shorter lifespans and less complex neuroanatomy than humans and postmortem studies in humans where sampling and other aspects of experimental design are difficult to control. These limitations underscore the need to examine basic mechanisms of brain aging in a more systematic manner in a species with a closer evolutionary affinity to humans.
Rhesus macaques are the best-studied nonhuman primate model species in medicine and neuroscience and recapitulate many aspects of human aging, including declines in bone mineral density and muscle mass, increases in circulating proinflammatory cytokines and increases in impairment of behavioral, sensory and cognitive function2. Critically, the individuals in our study lived under naturalistic conditions with minimal human interference before being humanely killed as part of necessary population control measures3. This allowed us to characterize aging in a natural cross-section of healthy adults, incorporate detailed behavioral and demographic information and sample high-quality tissues in a consistent manner. In doing so, we were able to experimentally control for autopsy biases4 and postmortem artifacts5 common to human studies.
Here, we leveraged a unique free-ranging rhesus macaque study system and profiled 36 animals (20 female and 16 male) over 15 cortical and subcortical brain regions (combined N = 527 samples) to quantify age-associated differences in cellular and molecular function across the brain and to test for associations with disease and the social environment. We also used single-nucleus RNA sequencing (RNA-seq) to profile cell-type heterogeneity in aging, concentrating on the dorsolateral prefrontal cortex (dlPFC) in a sample of 24 female animals. We identified thousands of genes showing age-associated transcriptomic changes, including roughly 1,000 that show highly consistent changes across sampled brain regions. We also found broad similarity in signatures of aging between humans and macaques and, drawing on detailed behavioral records, demonstrated that high social rank in female macaques is associated with decelerated transcriptomic aging in free-ranging macaques.
Results
A multiregion atlas of transcriptional aging in the brain
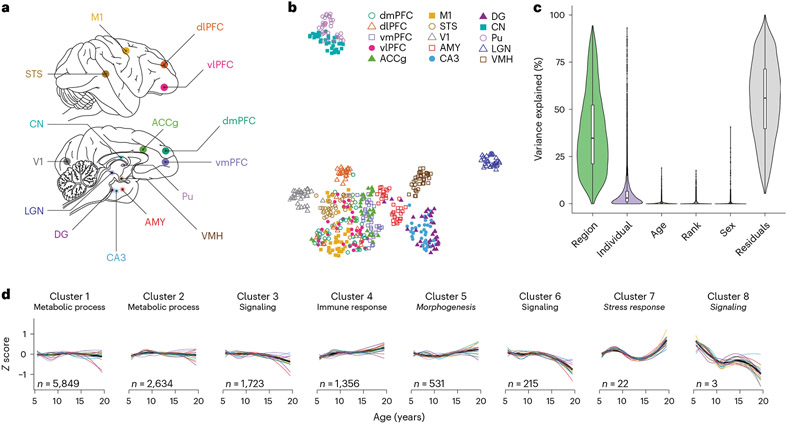
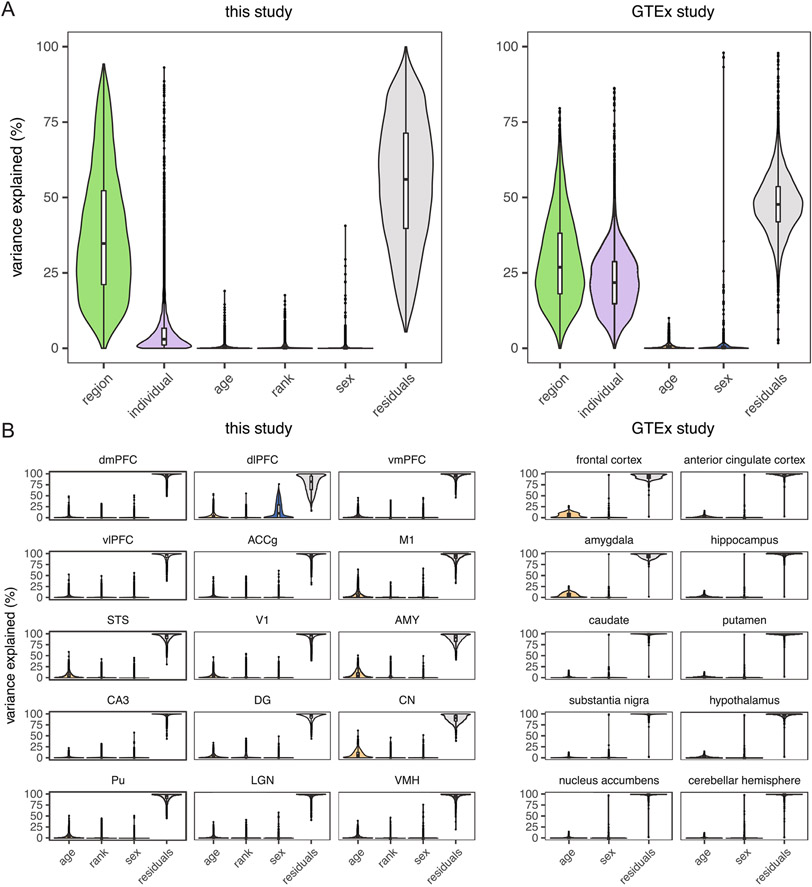
We first generated bulk tissue RNA-seq data from a total of 527 samples from 36 individuals (mean = 9.72 years old; range = 5.07–19.89 years; Supplementary Table 2) across 15 diverse brain regions (Fig. 1a, Extended Data Fig. 1 and Supplementary Table 1). Across our dataset, samples from the same broad anatomical unit (for example, cerebral cortex) were transcriptionally more similar to one another (Fig. 1b and Extended Data Fig. 2), and a median of 34.8% of transcriptional variance across genes was explained by brain region, the greatest proportion of any tested variable (Fig. 1c and Supplementary Fig. 3a). Biological variables such as age (median = 0.19%) and sex (median = 4.2 × 10−7%) explained much smaller proportions of variance in our global dataset. The explanatory power of age increased substantially when we examined each brain region alone, with age explaining the greatest proportion of transcriptional variance (0.6–6.4%) of all tested variables (Supplementary Fig. 3b). To benchmark these findings, we compared these values to estimates from our analysis of human brain transcriptomic data from the Genotype–Tissue Expression (GTEx) Consortium V8 dataset6, following a nearly identical pipeline (Methods). We found that age explained similar proportions of variance in human brain gene expression (median of 0.26% across 10 brain regions and 0.3–4.2% within regions; Extended Data Fig. 3).
Fig. 1 ∣. Experimental design and global expression patterns.
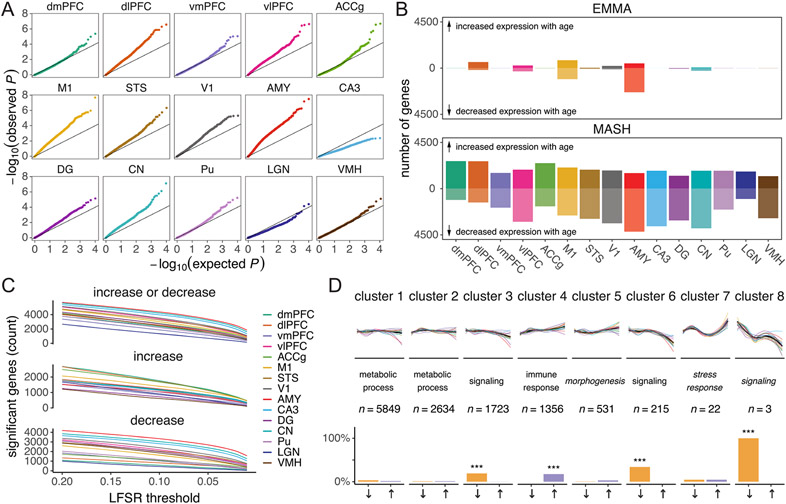
a, Brain regions sampled (N = 15 regions); dmPFC, dorsomedial PFC; dlPFC, dorsolateral PFC; vmPFC, ventromedial PFC; vlPFC, ventrolateral PFC; ACCg, anterior cingulate cortex gyrus; M1, primary motor cortex; STS, superior temporal sulcus; V1, primary visual cortex; AMY, amygdala; CA3, cornu ammonis 3; DG, dentate gyrus; CN, caudate nucleus; Pu, putamen; LGN, lateral geniculate nucleus; VMH, ventromedial hypothalamus. b, UMAP plot reveals the latent structure among bulk tissue RNA-seq libraries driven primarily by brain region, with distinct separation of the striatum, thalamus, cerebral cortex and other subcortical regions (hippocampus, AMY and hypothalamus). c, Age explains a relatively small percentage (median = 0.19%) of global variation in gene expression across the brain (N = 527 biologically independent samples). Box plots depict the median (center) and interquartile range (IQR; bounds of the box), with whiskers extending to either the maxima/minima or to the median±1.5× IQR, whichever is nearest. d, Clustering by age changes reveals eight clusters of genes with distinct trajectories. Within each cluster, expression was averaged across all genes and summarized separately by region (thin lines, colored by region) and combined across regions (thick black line). Clusters are labeled with high-level biological processes enriched within each cluster (see Supplementary Table 4 for detailed results), with italics indicating that the top processes did not pass a significance threshold (FDR < 0.2). The number of genes (n) assigned to each cluster is indicated.
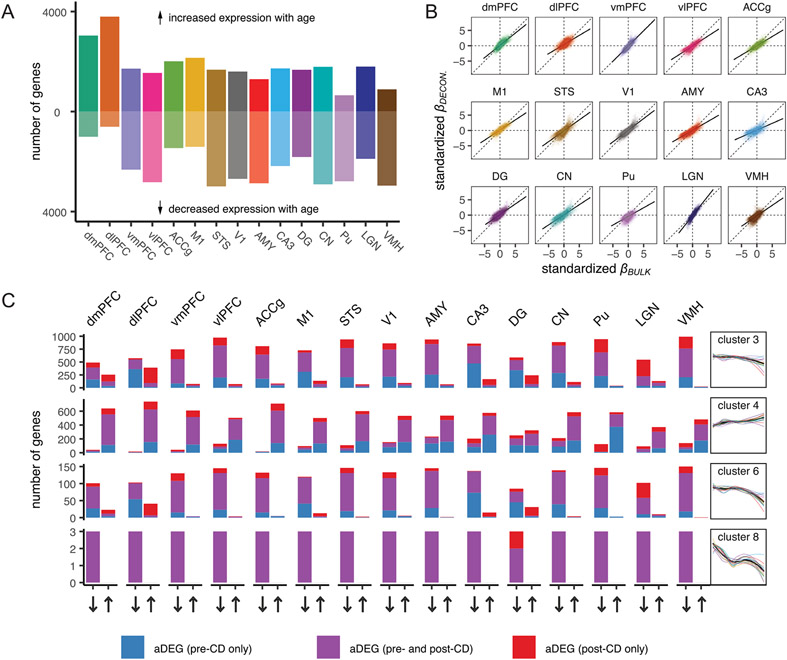
We next characterized non-linear aging trajectories across 12,332 detectibly expressed genes and clustered these trajectories into eight distinct transcriptional aging patterns (Fig. 1d). Diverse brain regions exhibited similar trajectories within each cluster, qualitatively similar to age-associated trajectories across organs in the mouse7, and were implicated in important biological processes (see Supplementary Table 4 for detailed enrichment results). Three clusters that decreased in expression with age (clusters 3, 6 and 8) were broadly involved in neural signaling and synaptogenesis. One cluster (cluster 4) that increased in expression with age was broadly enriched for genes involved in the immune response and the oxidation–reduction process, which may reflect increasing levels of inflammation and reactive oxygen species, respectively, that characterize aging8,9.
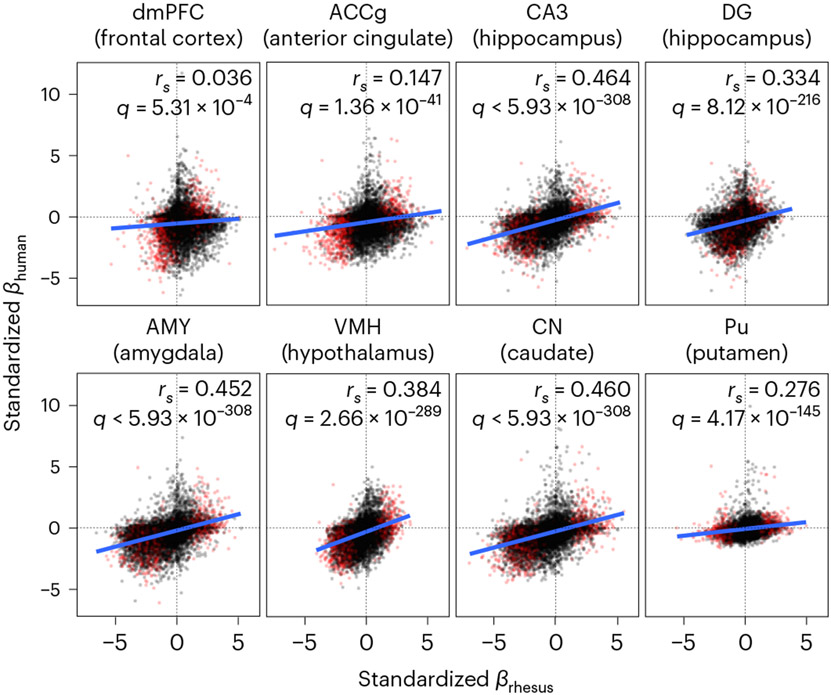
After identifying global transcriptional trajectories of aging across the brain, we next examined the effect of aging on the expression of individual genes within each of the 15 brain regions using linear mixed models controlling for genetic relatedness10, sex, dominance rank and technical covariates (Methods). Because our analyses were performed in parallel across 15 tissue types, we leveraged information across tissues to refine estimates of shared and unshared effects of age on expression11 and to estimate local false sign rates (LFSRs), which measure confidence in the directions of effect estimates while controlling for multiple tests12. Our age effect estimates were concordant with estimates from our analysis of human brain data (GTEx V8 (ref. 6); Methods), revealing strong parallels between macaque and human transcriptomic signatures of aging. We found significant positive correlations (Spearman’s ρ = 0.036–0.464; false discovery rate (FDR) < 0.001) between standardized age effect estimates of rhesus macaques and humans among one-to-one orthologous genes across eight overlapping brain regions (Supplementary Table 18), suggesting a high utility and relevance of macaques for modeling molecular aging in the human brain.
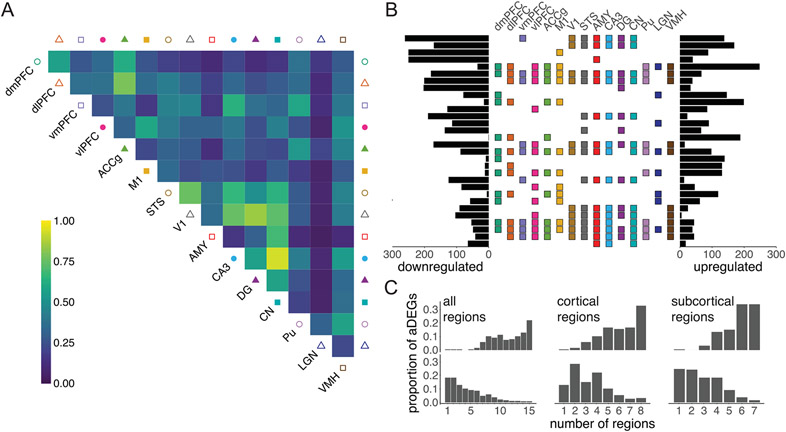
Overall, we identified 7,873 age differentially expressed genes (aDEGs), which exhibited significant differences in gene expression with age in at least one brain region (LFSR < 0.2; Fig. 2a), accounting for 63.8% of all measured genes. The region with the most aDEGs was the amygdala (AMY; 5,708 aDEGs), followed by the caudate nucleus (CN; 5,607 aDEGs) and cornu ammonis 3 of the hippocampus (CA3; 5,402 aDEGs), with strong majorities (67.8–73.4%) of these genes decreasing in expression with age. The relative rankings of brain regions according to the number of aDEGs were robust to LFSR thresholds (Extended Data Fig. 4c). These regions showing the broadest molecular changes with age are also among the first to be affected by age-associated neurodegenerative diseases13,14, suggesting that neurodegenerative disease pathologies in these regions may in part derive from cumulative changes occurring in these genes during normal aging. Our multiregion approach also allowed us to quantify the shared or specialized nature of the effects of age on the brain by distinguishing genes that are uniquely associated with age in one or a subset of regions from those that exhibit similar age-related changes across multiple regions (Fig. 2b and Extended Data Fig. 5). These classifications have important implications for understanding the molecular causes and consequences of neurological disorders of aging in a region-specific manner. Indeed, while the majority of aDEGs were shared among at least two regions, a sizable fraction (13.8%) exhibited directional changes that were unique to one brain region, suggesting that some aging mechanisms are region specific.
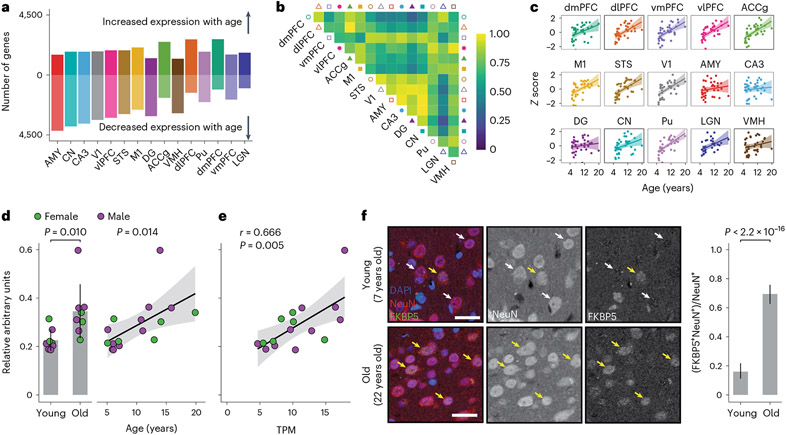
Fig. 2 ∣. Age broadly influences mean transcription levels across the brain.
a, Number of aDEGs across brain regions (LFSR < 0.2) ordered by number of genes. b, Proportion of aDEGs with shared signs between brain regions. For each pair of regions, genes are included if they were significant (LFSR < 0.2) in either region. c, The stress response gene FKBP5, considered a wbaDEG by our study, strongly increases in expression with age across nearly all brain regions. Error bands represent the 95% confidence interval of linear model predictions. d, Western blotting of bulk lysates from the dlPFC (N = 16 biologically independent samples) reveals an age-associated increase in FKBP5 protein expression normalized to GAPDH expression when partitioning animals into old versus young categories using a threshold of 10 years (1.53-fold increase; one-sided Student’s t-test, P = 0.010; left) and when analyzing continuous chronological ages (linear model including sex as a covariate, P = 0.014; right). Error bars represent the mean ± 1 s.d. Error bands represent the 95% confidence interval of linear model predictions. TPM, transcripts per million. e, Significant positive correlation between normalized FKBP5 gene expression (x axis) and normalized FKBP5 protein expression (y axis; two-sided Pearson’s correlation test, r = 0.666, P = 0.005). Error bands represent the 95% confidence interval of linear model predictions. f, Representative confocal images of the dlPFC (left) sampled from young (7 years old) and old (22 years old) captive macaques after immunostaining for FKBP5 (green), the pan-neuronal marker NeuN (red) and the nuclear marker DAPI (blue). After sampling 200 NeuN-labeled cells from each brain, we counted (right) 32 FKBP5+NeuN+ neurons (16.0%) in the young macaque compared to 139 FKBP5+NeuN+ neurons (69.5%) in the old macaque (one-sided Fisher’s exact test, P < 2.2 × 10−16); scale bar = 25 μm. Yellow arrowheads indicate FKBP5+NeuN+cells. White arrowheads indicate FKBP5− NeuN+cells. Points represent percentage estimates, and error bars represent 95% confidence intervals surrounding the estimate, which were estimated using the binomial distribution.
Another sizable fraction of aDEGs was robustly identified as significant in almost all brain regions, suggesting that these aDEGs play a universal role in aging across the brain (Supplementary Table 5). We identified 1,007 whole-brain aDEGs (wbaDEGs) that exhibited shared directional changes (LFSR < 0.2) in at least 13 of the 15 brain regions in this study, accounting for 12.8% of age-associated genes and 8.2% of all genes in this study. Among the most strongly supported wbaDEGs (LFSR < 0.005 in ≥13 regions; N = 13 genes) increasing in expression with age were the genes FKBP5 (Fig. 2c), a regulator of glucocorticoid receptor sensitivity whose increase has been linked to depression15, schizophrenia16 and AD pathology17, and SERPINA1, a risk marker for Parkinson’s disease18 and amyotrophic lateral sclerosis (ALS)19. One of the most strongly supported wbaDEGs decreasing in expression with age was ARPP21, which plays a protective role in the brain by inhibiting neural apoptosis and reducing inflammation of astrocytes20 and has also been linked to ALS neurodegeneration21.
To assess whether age-associated mRNA changes were linked to changes in protein expression22,23, we focused on one brain region, the dlPFC, and the gene FKBP5, which encodes the protein FKBP5 and was identified as a wbaDEG in this study and in our analysis of human brain data (GTEx V8 (ref. 6); β > 0 and LFSR < 0.05 in all tested regions; Methods). FKBP5 protein levels in the dlPFC increased with age (Fig. 2d, Supplementary Fig. 8 and Supplementary Table 22; linear model: P = 0.014; N = 16) and were significantly correlated with FKBP5 mRNA expression measured in the same individuals (Fig. 2e; Pearson’s r = 0.666, P = 0.005; N = 16). Using immunohistochemistry, we then assessed the cellular localization of FKBP5 expression differences in one young (7 years old) and one old (22 years old) captive macaque. We found a 4.34-fold increase in the fraction of FKBP5+NeuN+ double-labeled neurons (Fig. 2f; Fisher’s exact test, P < 0.001), indicating that increases in FKBP5 expression are attributable at least in part to changes involving neurons.
Biological processes and disease in the aging transcriptome
We found roughly twice as many wbaDEGs that decreased in expression with age relative to those that increased in expression with age (380 increasing wbaDEGs and 627 decreasing wbaDEGs). The age-increasing wbaDEGs were enriched for biological processes related to small-molecule biosynthesis, lipid metabolism and oxidationreduction (FDR < 0.05; Supplementary Fig. 5d and Supplementary Table 6). The age-decreasing wbaDEGs were involved in biological processes related to cell communication, synaptic signaling, neurogenesis, behavior and locomotion (FDR < 0.05; Supplementary Fig. 5d and Supplementary Table 6), suggesting that these genes may play roles in declines in neurological function with age. These age-decreasing wbaDEGs were also enriched for genes associated with mental disorders, including schizophrenia, autism, bipolar disorder and attention deficit hyperactivity disorder (FDR < 0.05; Supplementary Fig. 5d and Supplementary Table 8). Genes that are downregulated or upregulated in clinical cases of AD24 were also significantly enriched among age-decreasing (odds ratio (OR) = 1.67) and age-increasing wbaDEGs (OR = 1.95), respectively, indicating that changes in expression over the course of normal aging mirror those associated with AD and in shared directions. Parallel changes between age and AD were replicated in region-wise analyses for age-decreasing aDEGs in all 15 regions and age-increasing aDEGs in 9 of 15 regions (FDR < 0.05; Supplementary Table 10).
The ubiquitous age-associated declines in expression in wbaDEGs may result from reversals of expression patterns that increase in development and peak in early adulthood25, which is the lower age limit of animals in our study. Indeed, the promoters of negative wbaDEGs were enriched for predicted binding site motifs (FDR < 0.01; Supplementary Table 11) that included an overrepresentation of homeobox-family (FDR-adjusted P = 0.006) and Sox-family transcription factors (FDR-adjusted P = 0.006; Supplementary Table 12). Both homeobox and Sox transcription factors are key transcription factors involved in the development of a myriad of tissues, including the brain26,27, and Sox transcription factors are also involved in adult neurogenesis and brain homeostasis. Our findings thus suggest that broad age-related decreases in gene expression may reflect regulatory decoherence of developmental pathways that continue to function into later life (for example, continued roles in neuron maturation and identity) and/or declining neurogenesis28 and gliogenesis in older adults.
Shared signatures of aging between macaques and humans
Overall, we found broad similarities in age-related transcriptional changes between humans and macaques (Fig. 3), but this relationship was not isomorphic. We thus examined which aspects of the brain aging process were shared (or not) across species, indicating evolutionary conservatism (or divergence) and potentially highlighting the components of macaque neural aging that most closely model those in humans. Interestingly, out of 9 wbaDEGs in this study with the most robust support across regions (13 genes with LFSR < 0.005 in ≥13 regions; 9 of these have 1:1 orthologs in the human genome), 7 (77.8%) showed parallel age-associated changes across the vast majority of tested brain regions (LFSR < 0.2 in ≥90% of regions; Methods); these included FKBP5, SERPINA1 and ARPP21. We found that functional pathways exhibiting evolutionary conservatism (that is, functional groups of genes with highly concordant age effects in macaques and humans) outnumbered those exhibiting evolutionary divergence by over threefold in all overlapping regions (FDR < 0.05; Supplementary Tables 20 and 21). Among the pathways showing the greatest aging conservatism across regions were chemical synaptic transmission (shared across five regions), negative regulation of neurogenesis (three regions) and positive regulation of the proinflammatory cytokine tumor necrosis factor (three regions). Among the pathways showing the greatest aging divergence across regions was electron transport chain/oxidative phosphorylation (four regions). Interestingly, human neurodegenerative diseases, such as Parkinson’s disease (four regions), Huntington’s disease (three regions) and AD (one region), were associated with some of the most diverged gene sets, suggesting that, despite associations between macaque signatures of aging and human signatures of neurodegenerative disease reported here, normative trajectories of aging among some of these pathways may have diverged in the human lineage. These findings could potentially help explain the unique pathobiology of some neurodegenerative diseases in humans29-31 (Fig. 3).
Fig. 3 ∣. Parallel age-associated transcriptional signatures between macaques and humans.
Scatter plots depicting positive relationships between age effects estimated from macaques from this study (x axis) and humans from the GTEx study (y axis) in overlapping brain regions (GTEx regions are shown in parentheses). wbaDEGs, which show the most robust support for shared effects across macaque brain regions, are highlighted in red.
Age-dependent increases in transcriptional variability
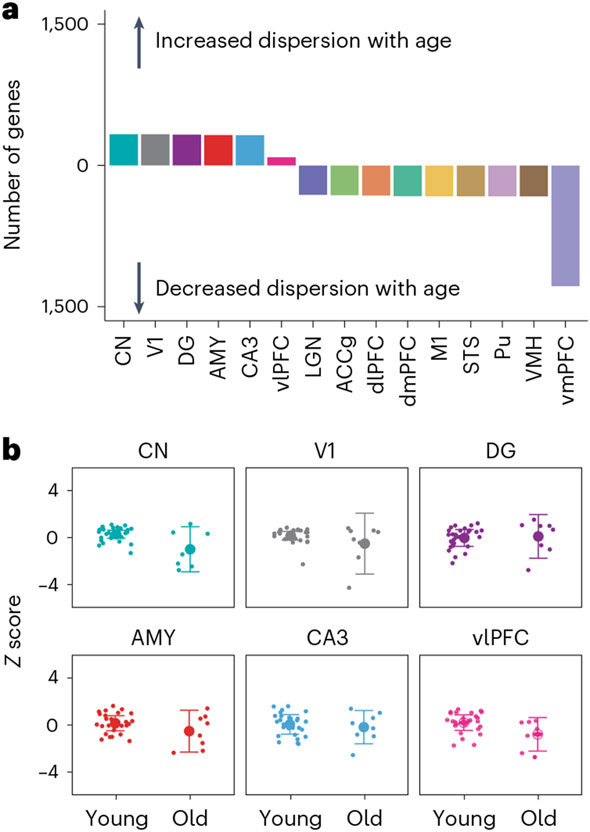
Aging and disease etiology are not solely characterized by monotonic changes in the mean values of biological phenotypes. Age-associated diseases can also be due to a decline in the control of molecular phenotypes, leading to increased variance and hence dysregulation in those phenotypes in older individuals32,33. To test this hypothesis, we used double generalized linear models (DGLMs) to jointly model the mean and dispersion of gene expression as a function of age and pooled information across tissues to refine estimates of shared effects among tissues (Fig. 4a and Methods). We focused primarily on genes with significantly increased variance of expression in older animals (Fig. 4b) to identify probable indicators of age-associated dysregulation (LFSR < 0.2). We found the largest number of these ‘age dysregulatory’ genes (N = 235) in a group of regions including the primary visual cortex (V1), AMY, CA3, dentate gyrus (DG) and CN, with a smaller number of genes (N = 86) detected in all of these regions and the ventrolateral PFC (vlPFC; Fig. 4a, Supplementary Fig. 7 and Supplementary Table 13). These genes were enriched for only two biological processes (FDR < 0.05), inflammatory response and axonemal dynein complex assembly (Supplementary Table 15), indicating that age-associated dysregulation may be strongest in inflammatory and dynein-mediated cell transport mechanisms in the brain, both of which have been implicated in the progression of AD34,35 and other neurodegenerative diseases36,37. Genes increasing in variance with age were also significantly enriched with genes differentially expressed in AD38 (Supplementary Table 17), consistent with age-associated dysregulation as a potential mechanism in AD pathogenesis.
Fig. 4 ∣. Age-associated changes in variance of transcription across the brain.
a, Number of genes with significant age-associated changes in dispersion of gene expression (LFSR < 0.2). b, Example of a gene (USP7) increasing in variance of expression with age in a group of brain regions, including CN, V1, DG, AMY, CA3 and vlPFC (N = 36 biologically independent animals). Animals were classified into young or old categories based on the midpoint of the sample age distribution. Data are presented as mean ± variance of gene expression.
Cell-type heterogeneity in signatures of aging
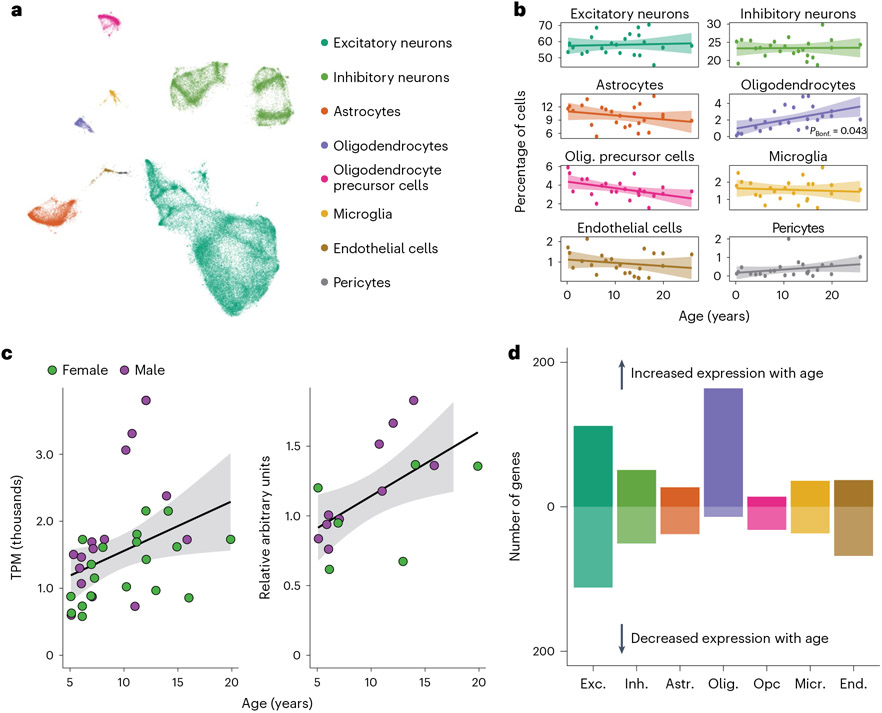
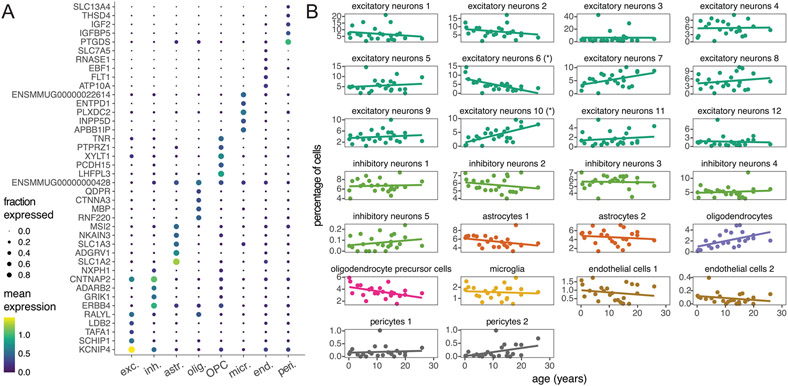
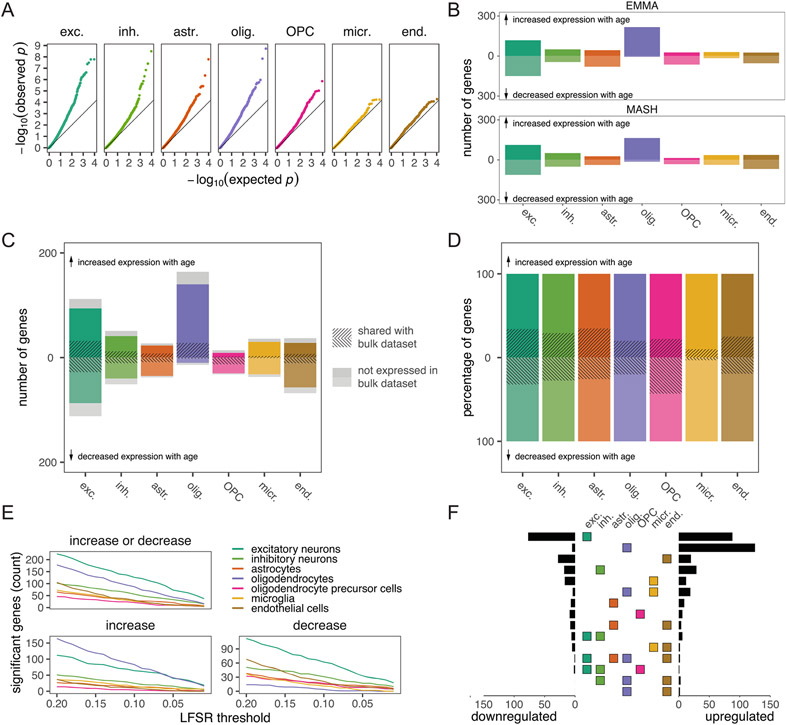
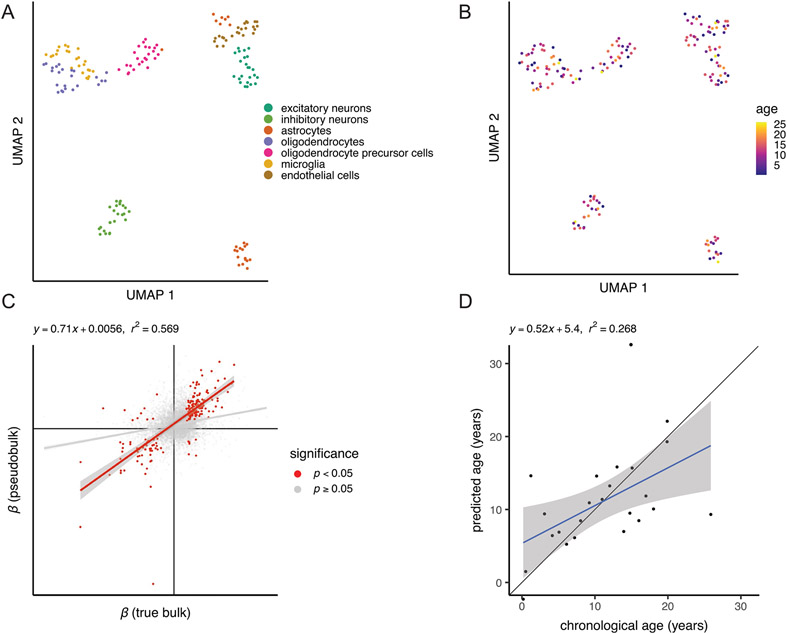
Bulk tissue gene expression data limit our ability to identify age-associated differences in gene expression in a cell-type-specific manner. To examine how aging in the brain varies among cells, we generated 71,863 single-nucleus RNA-seq transcriptomes from the dlPFC of 24 females spanning the full age range (Fig. 5a and Supplementary Table 3). We chose the dlPFC due to its homology between macaques and humans and its roles in cognitive decline and AD39-41. We first classified these nuclei into eight broad cell types based on marker gene expression (Supplementary Table 23) and harmonization with reference mouse and human brain single-cell RNA-seq data from the Allen Brain Map (Supplementary Fig. 11 and Supplementary Tables 24 and 25)42. Next, we used unsupervised clustering with the Leiden algorithm to identify a total of 26 distinct cell types and subtypes in the dlPFC (Supplementary Fig. 10). Oligodendrocytes were the only broad cell type that significantly changed in proportion with age (Fig. 5b and Supplementary Table 28). These cells increased in relative abundance (Bonferroni-adjusted P = 0.043), corroborating histological observations in rhesus macaques43. We further found that myelin basic protein (MBP), a canonical oligodendrocyte marker, increased in both bulk gene and protein expression in the dlPFC (Fig. 5c, Supplementary Fig. 8 and Supplementary Table 22), consistent with age-associated increases in oligodendrocyte density and myelination. Among the 26 cell subtypes, however, we identified a significant age-related increase (Bonferroni-adjusted P = 0.024) and decrease (Bonferroni-adjusted P = 0.014) in proportions of two subtypes of excitatory neurons (Extended Data Fig. 6 and Supplementary Table 29), which correspond most closely to layer 2 LINC00507/GLRA3 excitatory neurons and layer 2/layer3 RORB/RTKN2 excitatory neurons, respectively.
Fig. 5 ∣. Age alters both brain cell proportions and cell-type-specific expression.
a, UMAP of 71,763 single-nucleus transcriptomes from the dlPFC depicts broad neural cell classes. b, Oligodendrocytes are the only cell type showing a significant change in proportion with age (linear model, two sided; Bonferroni-corrected P = 0.043). Error bands represent the 95% confidence interval of linear model predictions; Bonf., Bonferroni; Olig, oligodendrocyte. c, Bulk analysis of the oligodendrocyte marker MBP from the dlPFC demonstrates strong age-associated increases in both normalized gene (left) and protein (right) expression, providing additional support for age-associated increases in oligodendrocyte density and myelination. Error bands represent the 95% confidence interval of linear model predictions. d, Number of aDEGs per cell type (LFSR < 0.2); Exc., excitatory neurons; Inh., inhibitory neurons; Astr., astrocytes; Opc, oligodendrocyte precursor cells; Micr., microglia; End., endothelial cells.
To investigate if age is associated with gene expression changes in specific cell types, we first combined our single-nucleus gene expression data into cell-type-specific pseudobulk counts per animal. We then modeled mean differences in cell-specific pseudobulk expression using a similar approach to that of our bulk tissue analysis. We identified 483 aDEGs in at least one cell type (LFSR < 0.2; Fig. 5d and Supplementary Table 31), and, of the subset analyzed in both our bulk and pseudobulk datasets (N = 390), 69.5% of cell-type aDEGS were also identified as aDEGS in at least one region of our bulk RNA-seq analysis. The lower number of aDEGs identified in our pseudobulk analyses relative to our bulk tissue analyses reflects decreased statistical power due to smaller sample size in terms of regions (N = 1 compared to 15 in the bulk dataset) and animals (N = 20 compared to 36). Notably, of the cell-type aDEGs overlapping with the bulk RNA-seq analysis, only 38.7% were also identified as aDEGs in the dlPFC. This large proportion of aDEGs newly identified by our pseudobulk analyses likely reflects cell-type-specific changes with age that are masked when measured in a heterogeneous tissue, as is typical for bulk RNA-seq. Consistent with this hypothesis, we found that the highest proportion of uniquely identified aDEGs was detected in microglia, one of the rarest cell populations (Extended Data Fig. 7d). We found that, while controlling for estimated cell-type proportions resulted in similar aDEG counts with highly correlated effect estimates (Extended Data Fig. 8a,b), an average of 38.6% (±9.1%) of aDEGs across regions did not meet our thresholds (LFSR < 0.2) to qualify as aDEGs following cell-type deconvolution (Extended Data Fig. 8c). Our results thus suggest that age-associated changes at the bulk tissue level are driven both by cellular shifts in expression and compositional changes in cell proportion.
We identified biological processes that were enriched among age-associated increases or decreases in cell-type-specific expression (nominal P < 0.001; Supplementary Table 32), indicating altered functions over the course of normal aging within cell populations. We found, for instance, that genes related to glutamatergic synaptic signaling are downregulated with age in excitatory neurons, as are genes regulating the modulation of excitatory postsynaptic potential in astrocytes. In oligodendrocytes, genes related to adenylate cyclase-activated G-protein-coupled receptor signaling, which induces oligodendrocyte differentiation44, were downregulated. In microglia, genes related to regulation of DNA damage checkpoint, and immune response activation via cell surface receptor signaling were downregulated, while cellular response to interferon-γ was upregulated, consistent with age- and disease-associated priming of microglia to a proinflammatory state45,46.
Aging is associated with variation in the social environment
While health declines are an inevitable feature of aging, evidence in humans and other social species suggests that variability in the risk, onset and progression of age-related morbidities47,48 is explained in part by variation in social adversity49. In female macaques, for instance, low dominance rank, an index of social adversity, is associated with increased mortality50, and its effects on peripheral blood transcriptomes recapitulate the transcriptional signatures of aging51. We next aimed to determine whether social adversity is associated with accelerated transcriptomic signatures of aging in the macaque brain.
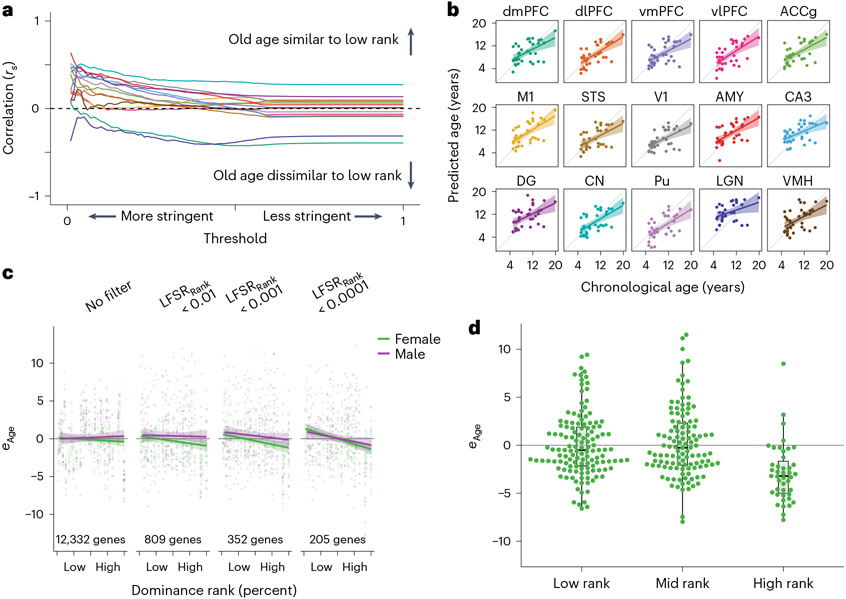
We first tested if low dominance rank recapitulated the effects of aging on the brain transcriptome. Using behavioral observations of dyadic agonistic interactions, we calculated each individual’s dominance rank as the percentage of same-sex groupmates outranked52 (Methods). We found that, even without imposing any precision-based filters on effect estimates, 7 of 15 brain regions in our analysis (dlPFC, anterior cingulate cortex gyrus, primary motor cortex (M1), V1, AMY, DG and CN) exhibited significant positive correlations between aging and social adversity (low dominance rank) effects for all genes in our analysis (Bonferroni-adjusted P < 0.05). These shared effects strengthened across all 15 regions when we excluded genes failing increasingly stringent significance thresholds (Fig. 6a and Supplementary Fig. 13). While thresholding genes by nature focuses on stronger effects, both positive and negative, it should only increase correlations in the presence of true shared effects between older ages and lower dominance ranks. Our results thus demonstrate that the gene expression signatures of aging and social adversity parallel one another.
Fig. 6 ∣. Social status signatures of the aging transcriptome.
a, Correlations between age and dominance rank effects robustly increase at more stringent statistical thresholds, indicating similarities between older ages and lower dominance rank. For each threshold, effect estimates were retained when LFSRAge and LFSRRank both passed the threshold on the x axis. Lines correspond with brain regions and follow colors in b. b, An age prediction model robustly predicts chronological age across brain tissue types. All predictions were run using wbaDEG coefficients. Error bands represent the 95% confidence interval of linear model predictions. Prediction results using elastic net (‘glmnet’) models showed similar results (Supplementary Fig. 14). c, Age prediction models filtered to genes with increasingly well-supported dominance rank effects on gene expression reveal stronger support for dominance rank effects on relative transcriptomic age (eAge). Samples from all individuals and brain regions are plotted together. Error bands represent the 95% confidence interval of linear model predictions. d, Comparison of categorical dominance ranks reveals that dominance rank effects on female aging are driven primarily by younger relative ages in high-ranking females. Samples from all individuals and brain regions are plotted together (N = 291 biologically independent samples). Results are shown for predictions filtered first to genes with LFSRRank < 0.001 but were similar across multiple thresholds (Supplementary Fig. 15 and Supplementary Table 37). Box plots depict the median (center) and IQR (bounds of box), with whiskers extending to either the maxima/minima or to the median ± 1.5× IQR, whichever is nearest.
We next identified functions most associated with shared signatures of aging and social adversity across brain regions by repeating our analysis across biological processes and pathways (FDR < 0.05; Supplementary Tables 33 and 34). We found that among the pathways showing the most robust sharing across regions were the proteasome (four regions) and oxidative phosphorylation (three regions) as well as neurodegenerative diseases, including Parkinson’s disease (three regions), Huntington’s disease (three regions), AD (three regions) and ALS (two regions). The brain regions with the strongest statistical support among neurodegenerative disease gene sets were the M1, DG and CN, all of which are associated with motor or cognitive function and have previously been linked to AD and other neurodegenerative diseases53-55. Our results suggest that, while neurodegeneration pathways in humans are relatively divergent from macaques in their aging profiles in some regions, they nevertheless exhibit strong overlap with social adversity, paralleling epidemiological links in humans between social adversity and neurodegenerative diseases56-58.
We next tested if the social adversity of low dominance rank is associated with accelerated aging using a broad measure of brain transcriptomic aging. To do so, we developed two age prediction models, one based on model coefficients of wbaDEGs (LFSR < 0.005 in ≥13 brain regions) averaged over all brain regions and the other based on elastic net regression. Both models predicted age in held-out samples with significant accuracy (wbaDEGs: P = 1.08 × 10−39; elastic net: P = 7.66 × 10−32; Fig. 6b, Supplementary Fig. 14 and Supplementary Table 35). From these transcriptional age predictions, we characterized individual samples as either younger or older than their chronological age by calculating a ‘relative transcriptomic age’ (eAge), which we defined as the residuals of the linear regression relating chronological age and predicted age. We did not find support for the prediction that low dominance rank is associated with positive eAge using either our wbaDEGs (P = 0.407) or elastic net regression models (P = 0.165). We hypothesized, however, that a true effect of dominance rank on a subset of age-associated genes could be masked if a substantial fraction of age-predictive genes selected by our models are only weakly linked to social environmental effects on aging.
To test this hypothesis, we iteratively filtered our dataset to exclude genes with relatively weak dominance rank effects and reran model site selection and age predictions. When we focused on genes that were more strongly associated with dominance rank (LFSRRank < 0.0001), we found that low-ranking individuals had transcriptionally older brains as measured by eAge This effect was robust to our choice of thresholds (Supplementary Fig. 16 and Methods) and was driven by females (Bonferroni-adjusted P = 0.033; Fig. 6c, Supplementary Fig. 15 and Supplementary Table 36), which may reflect the relative lifetime stability of dominance rank in female macaques, who inherit their ranks from their mother and, for the most part, remain at those ranks throughout their lives. This contrasts from the pattern in males, who disperse from their natal groups and typically enter their new groups at the bottom of the hierarchy before rising in rank as their tenure in the new group lengthens59. The effect of rank on transcriptional age was particularly driven by decelerated age phenotypes in high-ranking females (Fig. 6d, Supplementary Fig. 15 and Supplementary Table 37), suggesting that associations between increased rank and slower aging are not expressed linearly along the social hierarchy but instead are specific to females with the highest ranks, who may reap advantages including increased access to resources, more predictable environments and decreased harassment from groupmates.
Discussion
Here, we profiled transcriptional aging across 15 cortical and subcortical brain regions in a representative cross-section of healthy adult rhesus macaques living in naturalistic conditions (N = 36 animals and N = 527 samples). We complemented these data with single-nucleus transcriptional measures in the dlPFC in 24 females spanning the macaque lifespan (N = 71,863 nuclei). We found evidence for both tissue-specific and shared signatures of aging and cell-type-specific signatures of aging. Notably, the transcriptional signature of macaque brain aging broadly recapitulates major features of human brain aging. We also found that age-associated changes in mean expression or variability of expression involve genes associated with neurodegenerative diseases, including AD. Together, our findings provide a detailed transcriptional resource cataloging brain aging in a model nonhuman primate in naturalistic environments.
These atlases and findings provide valuable targets for future studies in a tractable, clinically important model of human health and aging. Moreover, we found evidence for a putative molecular link between social adversity and accelerated aging in the brain, complementing similar findings from the peripheral blood immune system in humans51,60-62. This link potentially has a causal explanation; the chronic stress of social adversity, for instance, has been proposed to accelerate aging by downregulating glucocorticoid receptor sensitivity and promoting chronic inflammation63. In support of this hypothesis, social manipulation experiments in rhesus macaques have demonstrated a causal relationship between low dominance rank, attenuated glucocorticoid sensitivity and altered immune regulation in peripheral blood mononuclear cells64,65. Given the correlative nature of our study, we are at present unable to determine if social adversity accelerates aging in the brain or if the correlation is explained by other factors; for instance, both aging and dominance rank could be correlated with a third variable. Given that female macaque ranks are inherited matrilineally and remain stable throughout life, it is more plausible that the benefits of high social status in females lead to slower aging than vice versa. Our work demonstrates that the aging transcriptome may be influenced by variation in naturalistic environments and underscores the importance of the social environment as a modifier of aging and health.
Methods
Study population and sample
The rhesus macaques of Cayo Santiago were introduced to the island of Cayo Santiago (Extended Data Fig. 1a), PR, in 1938 and have been the subjects of nearly continuous study ever since66. Apart from being provisioned with commercial feed and subject to occasional capture and release, macaques on Cayo Santiago live in naturalistic circumstances largely free from human interference. Under these conditions, they form social groups and relationships that mirror those of rhesus macaques in the wild. Female macaques on Cayo Santiago, for instance, form stable linear dominance hierarchies in which dominance ranks are inherited genealogically through matrilines67, while male macaques form unstable dominance hierarchies by which dispersing males obtain the lowest dominance ranks after immigrating to new social groups and increase in rank over their tenures59. The macaques of Cayo Santiago also maintain an outbred population structure despite decades of isolation68.
The rhesus macaques on Cayo Santiago are maintained by the Caribbean Primate Research Center (CPRC). The average annual growth rate of the Cayo Santiago population far surpasses that of rhesus macaques in the wild, making capture and removal of animals necessary as a means of population control3. In 2016, the CPRC began selectively removing entire social groups as part of an updated population management strategy. Concurrently, the Cayo Biobank Research Unit (CBRU) was created to maximize the research potential from these activities by collecting and archiving postmortem tissues and data from animals removed by CPRC. The data in this manuscript are derived from the CBRU’s archive. All procedures related to capture, removal and euthanasia were conducted by CPRC in accordance with protocols approved by the animal use committee of the University of Puerto Rico (protocol number 338300).
Because entire social groups (one social group per season) were removed over the course of a 3-month season, there were no trapping-related selection biases among individuals (for example, bias in health or age status).
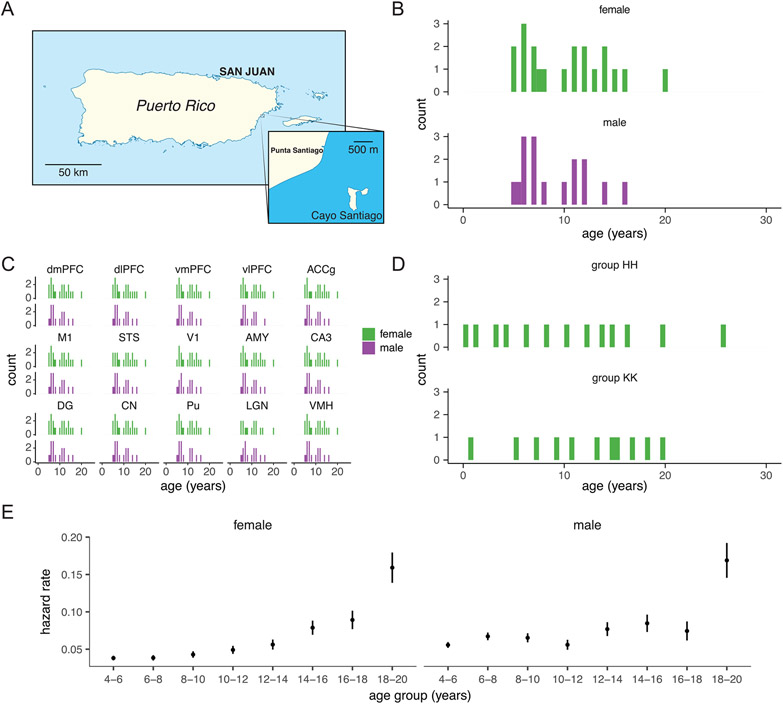
Bulk RNA-seq samples were collected from 36 animals (20 females and 16 males) who were removed by CPRC in 2016. All individuals were from a single social group (HH) and were representative of the natural adult (after sexual maturation) age distribution in this population (median = 9.18 years and range = 5.08–19.89; Extended Data Fig. 1b,c and Supplementary Table 2). Notably, the age distribution of free-ranging macaques on Cayo Santiago skews far younger than that of captive rhesus macaques2, the latter of which are subject to husbandry practices that have substantially prolonged rhesus macaque lifespans in laboratory environments69. Our sample is representative of ages among removed adults and, given the exhaustive removal of entire social groups, therefore captures the true distribution of adult ages of free-ranging macaques on Cayo Santiago.
Given that median life expectancy in captive rhesus macaques is reported to be around 25 years (ref. 70), the free-ranging macaques of Cayo Santiago would not be considered old by laboratory standards. Of 11,659 animals monitored in the history of the Cayo Santiago colony (as of March 2021), only 33 (0.28%) survived to 25 years, underscoring the compressed lifespan of naturalistic macaques. Cayo Santiago macaques, however, show several indicators of accelerated aging, including a geriatric appearance attained at younger ages71, premature reproductive senescence72 and accelerated rates of degenerative joint disease and restricted mobility73. Moreover, hazard rates from published demographic models2 demonstrate that Cayo Santiago animals experience elevated risks of mortality well within the age range of our sample (Extended Data Fig. 1e). These findings are consistent with theoretical and empirical evidence that, despite greater exposure to extrinsic mortality, senescence is widespread in wild populations74, particularly among primates75. Given our representative sampling design, the evidence indicates that our sample captures normative trajectories of aging in this naturalistic population of macaques.
Single-nucleus RNA-seq samples were collected from 24 animals (all female) from Cayo Santiago who were removed by CPRC in 2016 and 2018. Individuals were from two social groups (HH and KK) and were selected to uniformly sample the natural age distribution and to minimize social group effects by sampling evenly between groups (median = 12.01 years and range = 0.18–25.88; Extended Data Fig. 1d and Supplementary Table 3). Seven individuals from social group HH were sampled in both our bulk and single-nucleus RNA-seq datasets.
Behavioral data collection
We collected behavioral data from all animals for this study in the 3 months before removal. Methods for behavioral data collection as well as dominance rank inference in this population for these sampling years are described by Testard et al.52. We followed Testard et al. in creating two measures of dominance rank: (1) percent dominance ranks, calculated as the percentage of same-sex groupmates that an individual outranks, and (2) categorical dominance ranks, calculated by classifying animals as high (rank ≥ 80%), mid (50% < rank < 80%) or low ranking (rank < 50%) based on their percent dominance ranks. We modeled categorical dominance ranks as an ordinal variable for all differential expression analyses using the ordered factor class in R76. We modeled percent dominance ranks as continuous variables.
Sample collection
Brain collection procedures and, crucially, the interval between euthanasia and sample cryopreservation were standardized across all animals to minimize postmortem artifacts in the data. Following veterinary euthanasia, a necropsy was immediately performed during which the brain was perfused by transcardial perfusion with sterile saline. The brain was then removed from the cranium and sagittally sectioned into left and right hemispheres. The left hemisphere was fixed in 10% formalin and not used for this study77. The right hemisphere was further sectioned before cryopreservation. First, the cerebellum was separated from the cerebrum. The cerebrum was then placed on a custom mold and sectioned coronally into 11 approximately 0.5-cm-thick slices. All 12 slices (including the cerebellar hemisphere) were individually stored in Whirl-pak bags and flash-frozen in a dry ice/ethanol bath immediately before storage in ultralow (−80 °C) freezers. The interval between euthanasia and storage of frozen tissue averaged 51.09 min, with a standard deviation of 5.84.
The brain regions in this study (Fig. 1a and Supplementary Table 1) were dissected under sterile, RNase-free conditions on dry ice to minimize RNA degradation. Briefly, anatomical regions were identified using neuroanatomical landmarks78. For bulk RNA-seq analysis, six 1-mm tissue biopsies were then collected by biopsy punch (Integra, 3331AAP/25) and transferred to a frozen 2.0-ml microcentrifuge tube. For single-nucleus RNA-seq analysis, six 2-mm tissue biopsies were collected from the dlPFC by biopsy punch (Integra, 3331P/25) and transferred to a frozen 1.5-ml microcentrifuge tube.
Bulk RNA-seq data generation and quality control
RNA extraction.
Total RNA was extracted using a TRIzol–chloroform gradient followed by column isolation of RNA using the Quick-RNA Microprep kit (Zymo Research). Briefly, 1 ml of TRIzol (Invitrogen) was added to each 2-ml sample tube, along with a sterile 5-mm stainless steel grinding bead (QIAGEN, 69989). Samples were then homogenized using the Tissuelyser II system (QIAGEN) at 20 Hz in two sets of 2 min each with rotation in between, and the lysate was transferred to a new tube. Chloroform (200 μl) was added to each sample, which was then vortexed at full speed for 15 s and incubated at room temperature for 2 min. The sample was then centrifuged at 12,000g for 15 min at 4 °C.
After centrifugation, the aqueous layer was transferred to a new tube and mixed with 600 μl of 70% ethanol to precipitate RNA. The resulting solution was transferred to Quick-RNA Microprep columns (Zymo Research). The remainder of the protocol followed manufacturer’s instructions for the Quick-RNA Microprep kit. Total RNA was eluted in 60 μl of RNase-free water. RNA yield was quantified by Qubit 3 using the Qubit RNA HS assay kit (Invitrogen), and RNA quality was estimated by electrophoresis using either the TapeStation High-Sensitivity RNA ScreenTape (Agilent) or the Fragment Analyzer HS RNA kit (AATI/Agilent). RNA quality metrics from the TapeStation (RINe) and Fragment Analyzer (RQN) systems were standardized by applying corrections from previously published comparisons of RNA quality metrics79 (Supplementary Table 2).
Library preparation.
We prepared bulk RNA-seq libraries using the NEBNext Ultra II RNA library prep kit for Illumina (New England Biolabs, E7770L) with an initial mRNA isolation step using the poly(A) mRNA magnetic isolation module (New England Biolabs, E7490L). We followed the manufacturer’s instructions and guidelines for library preparation. We used 200 ng of starting RNA and set the enzymatic fragmentation time (10 min), first-strand synthesis incubation time (50 min at 42 °C) and bead cleanup volume ratios for a target insert size of 400 base pairs (bp). Libraries were amplified using 10 cycles of PCR and eluted in a final volume of 20 μl of RNase-free water. Libraries were quantified by Qubit 3 using the Qubit DNA HS assay kit (Invitrogen), and library quality was assessed using the Fragment Analyzer HS NGS kit (AATI/Agilent). Libraries were pooled in equimolar quantities before sequencing.
Sequencing.
Libraries were sequenced on Illumina NovaSeq S2 or S4 flow cells using 2 × 50 bp (S2) or 2 × 101 bp (S4) sequencing at the University of Washington Northwest Genomics Center core sequencing facility.
Sequence data analysis and quality control.
Gene expression counts were quantified from RNA-seq reads using the pseudoalignment method implemented in kallisto80. Pseudoalignment was performed using the most recent rhesus macaque reference genome (Mmul_10)81. Estimated counts and transcripts per million (TPM) values were then imported into R v4.0.2 (ref. 76) and summarized to genes using the tximport package82. We then transformed our estimated counts into log2 (count) values using voom from the limma package83 and removed all lowly expressed genes with an average TPM of <10 in all regions.
For exploratory data visualization, we removed technical covariates (batch, RNA integrity and number of mapped reads) from the log2 (count) expression values by running a linear model including the aforementioned technical covariates along with age, sex and dominance rank. We then extracted partial residuals from this model to produce a corrected expression matrix that retains the effects of biological covariates while eliminating the effects of technical variation. We visualized latent relationships among our samples in two dimensions by performing a uniform manifold approximation and projection (UMAP) analysis (n_neighbors = 50 and min_dist = 0.5 (ref.84); Fig. 1b and Supplementary Figs. 1 and 2). We also used hierarchical clustering to explore relationships among brain regions in our analysis (Extended Data Fig. 2). First, we z scored expression within each gene across all 527 samples from our corrected expression matrix. Next, we calculated mean z scores for each gene within each region and performed hierarchical clustering of Euclidean distances using the complete method. To visualize uncertainty in the clustering, we performed 1,000 bootstrap replicates in which we resampled with replacement libraries within each region and repeated mean z-score calculations and hierarchical clustering as described above for each replicate. Dendrograms including bootstrap replicates were visualized using the densiTree function from the phangorn package85.
To explore the contributions of technical (that is, batch, read counts and RNA integrity) and biological covariates (that is, brain region, age, sex, dominance rank and individual identity) on global gene expression, we performed variance partitioning on our voom-normalized global gene expression matrix using the variancePartition R package (Supplementary Fig. 3a). We also repeated variance partitioning on our corrected expression matrix (that is, with technical covariates regressed out) for both the combined dataset (Fig. 1c) and for each region separately (Supplementary Fig. 3b).
To explore effects of age on global gene expression patterns, we performed a principal components analysis for each region on Pearson’s correlations among samples, thereby standardizing variables across our analysis. We regressed the resulting principal components against age and focused on significant age-related differences in principal components explaining >99% of the total variance in gene expression (Supplementary Fig. 4).
All exploratory analyses were performed in R.
Genotyping.
We genotyped individuals using the RNA-seq data to estimate genetic relatedness among individuals. For each sample, we mapped reads to the rhesus macaque reference genome using the splice-aware algorithm STAR86 to generate alignment files in BAM format. We then pooled mapped reads for each individual across all brain regions using SAMtools87 to maximize read depth for each individual. We used the Genome Analysis Toolkit (GATK)88,89 to mark duplicates (MarkDuplicates), split reads spanning splice events (SplitNCigar-Reads) and recalibrate base quality scores (BaseRecalibrator and Apply-BQSR). We then called variants using GATK HaplotypeCaller with a standard minimum confidence threshold for calling of 20.0 and filtered variants using GATK VariantFiltration with a window size of 35, a cluster size of 3 and the filters FS > 30.0 and QD < 2.0. We removed variants that failed these filters and further filtered single nucleotide variants using VCFtools90 to a minimum distance of 100 kilobases (kb) between sites, a minor allele frequency of 0.3 and a minimum completeness of 0.9 (no more than 10% missing data) to generate the final genotypes.
We used lcMLkin91 to compute a relatedness matrix among individuals from the filtered and thinned variants using Phred-scale genotype likelihoods (PL) denoted in the VCF file.
Estimation of age trajectories from bulk gene expression
We calculated age trajectories from our bulk gene expression dataset using an approach based on that of Schaum et al.7. For this analysis, we used the log2 (count) gene expression matrix after removing technical effects described earlier and calculated mean z scores separately for each brain region. To estimate global age trajectories across the 15 sampled brain regions in our study, we assigned a single expression value for each animal for each gene, calculated as the median z score across all sampled brain regions. We then modeled the assigned median z scores by fitting a LOESS model with age as the predictor. We then used the LOESS model to predict gene expression for age values spanning our sample age distribution in 0.5-year intervals (5.5–19.5 years). We next performed hierarchical clustering of Euclidean distances using the complete method on the resulting age trajectory matrix and assigned genes to k = 8 clusters (Fig. 1d). We determined optimal k based on visualization of the clusters.
For each cluster, we tested for enriched biological processes by performing a Gene Ontology (GO) enrichment analysis using topGO92,93, which corrects for the correlated nature of the underlying GO graph structure. We used GO annotations from Ensembl (release 101) obtained using the biomaRt package94 and limited GO terms to the biological process (BP) ontology and eliminated terms with fewer than 10 associated genes (nodeSize = 10). We then implemented hypergeometric tests (statistic = ‘fisher’) using the ‘parentchild’ algorithm to identify overrepresented GO processes. We calculated q values using a Benjamini–Hochberg correction95 to correct for multiple hypothesis testing (Supplementary Table 4).
Estimation of linear age effects on mean bulk gene expression
Linear mixed modeling.
For each region, we modeled gene expression values using efficient mixed model association (EMMA) models10,96 to control for relatedness among individuals in our dataset (Extended Data Fig. 4). The EMMA algorithm fits a linear mixed model with population structure or relatedness included as a random effect. We ran EMMA models separately for each region.
For each region, we first excluded all genes previously identified as having a mean TPM of <10 in that region. We then modeled log2 (count) values with sex, age, dominance rank, RNA integrity scores, mapped read counts and library batches included as fixed effects. We also included a k (relatedness) matrix estimated using lcMLkin as described earlier as well as a z matrix for assigning libraries to genotypes. EMMA models were implemented in the EMMREML package in R.
Multivariate adaptive shrinkage (MASH).
To integrate information across brain regions, increase statistical power and refine our understanding of shared and region-specific effects, we used MASH11 implemented in the mashr package to adjust β estimates (that is, the effect sizes). MASH pools information across conditions and estimates a data-driven covariance matrix to improve power and to refine effect size estimates across conditions. As input, we used β estimates and standard errors of the β estimates from the age term from our EMMA models across all 15 brain regions in our analysis. Where values were missing due to our region-specific TPM filters, we set β to 0 and the standard error of β to 1,000 following recommendations of the mashr authors and others97.
We first ran a simple condition-by-condition analysis in mashr to identify an initial set of genes passing a significance threshold of 0.05 (the ‘strong’ set). We then randomly sampled half of all expressed genes to produce a null gene set (the ‘random’ set). We estimated data-driven covariance matrices from the ‘strong’ set of genes using the principal components analysis and extreme deconvolution methods implemented in mashr. We then ran a MASH model on the ‘random’ set of genes with the two covariance matrices estimated above as inputs to fit a mixture model. We ran a final MASH model on the full set of genes using the previously learned mixture model as input. We extracted posterior mean β estimates, posterior standard deviations of β and LFSRs12 from the MASH model. We considered genes with LFSR < 0.2 to be aDEGs (Fig. 2a and Extended Data Fig. 4).
Gene set enrichment.
We tested for enrichment of biological processes and diseases associated with age effects detected in (1) each region separately and (2) age effects universally shared across brain regions. We used a common threshold (LFSR < 0.2) but otherwise different significance criteria for the two approaches. For the first approach (region-by-region aDEGs), we considered genes to be significant for each region if they passed our threshold in that region. For the second approach (wbaDEGs), we considered genes to be universally significant across regions if they passed our threshold and shared the sign of their effect sizes in at least 13 of the 15 brain regions sampled in this study (Supplementary Table 5). We performed enrichment tests separately for each region for the first approach and once for the entire brain for our second approach. For all tests, we separately tested for enriched categories associated with positive β, indicating increased expression with age, and associated with negative β, indicating decreased expression with age.
We used topGO92,93 to test for age-related enrichment of biological processes using the two approaches described above. As with our trajectory analysis described earlier, we used Ensembl GO annotations obtained from biomaRt94 and limited GO terms to the biological process ontology and eliminated terms with fewer than 10 associated genes (nodeSize = 10). We tested for significance using hypergeometric tests (statistic = fisher) using the parentchild algorithm to identify overrepresented GO processes and calculated ORs by dividing the number of significant genes by the number of expected genes calculated by topGO (Supplementary Fig. 5a,d and Supplementary Tables 6 and 7). Notably, while P values are corrected for the GO graph structure by topGO, ORs are not.
We used gene–disease associations from the DISEASES database98 to test for age-related enrichment of diseases, again in region-by-region aDEGs and wbaDEGs. We used protein names and Ensembl identifiers along with orthology information in biomaRt94 to link disease associations to one-to-one orthologous genes in the rhesus macaque genome. We retained all non-redundant gene–disease associations and ran Fisher’s exact tests to test for enrichment in R (Supplementary Fig. 5b,d and Supplementary Tables 8 and 9).
Finally, we tested for age-related enrichment of genes that are differentially expressed in individuals with AD. We used previously reported DEGs from a meta-analysis of three RNA-seq studies38 (the MSBB99, ROSMAP100 and MayoRNAseq101 studies) obtained from the AMP-AD Knowledge Portal24 and encompassing seven brain regions, the cerebellum, temporal cortex, frontal pole, inferior frontal gyrus, parahippocampal gyrus, superior temporal gyrus and dlPFC. We used orthology information in biomaRt94 to link AD DEGs to one-to-one orthologous genes in the rhesus macaque genome. We then used a Fisher’s exact test to test for an overrepresentation of AD DEGs among region-wise aDEGs and wbaDEGs. We separately tested for overrepresentation of AD DEGs with positive or negative estimates among aDEGs/wbaDEGs of the same sign (Supplementary Table 10).
For all enrichment analyses, we adjusted P values using a Benjamini-Hochberg correction95.
Transcription factor motif enrichment.
We used HOMER v4.11 (ref. 102) to test for enrichment of vertebrate transcription factor-binding site motifs in the promoters of aDEGs. We used loadGenome.pl to add the latest version of the rhesus macaque genome (Mmul_10)81 and used findMotifs.pl to find motifs in the regions ±2 kb from transcription start sites of genes. For this analysis, we considered genes to be age associated if they passed an LFSR threshold of 0.05 in at least one brain region. We ran HOMER separately for genes that increased in expression with age and genes that decreased in expression with age (note that there is a small degree of overlap between these two sets due to the possibility that some genes increase significantly with age in some regions and decrease significantly with age in other regions). For each analysis, we set as the background all non-overlapping genes that passed our initial TPM filters in at least one brain region and considered motifs enriched if they passed a threshold of FDR < 0.01 (Supplementary Fig. 5c and Supplementary Table 11).
We next tested for an overrepresentation of age-associated transcription factor-binding motifs belonging to particular transcription factor families. We restricted this analysis to transcription factor families with at least 10 associated motifs in our dataset and, as with before, considered motifs enriched if they passed a threshold of FDR < 0.01. We tested for enrichment using one-sided Fisher’s exact tests and adjusted P values using a Benjamini–Hochberg correction95 (Supplementary Table 12).
Estimating age effects on variance of gene expression
We tested the prediction that age-related declines in gene regulatory mechanisms could result in increased variance of gene expression in older animals. To model age-related changes in variance, we ran DGLMs using the dglm package103 in R. DGLMs are a two-stage model in which the means and variance are modeled, respectively, by GLMs. The dispersion is typically modeled with a gamma error distribution. We ran DGLMs using the same covariates as our EMMA model. We modeled means using a Gaussian error distribution and modeled the dispersion using a gamma distribution with a log link function and setting age as the predictor.
Because each brain region was modeled separately, we used a similar MASH-based strategy to pool information across tissues and to refine our effect estimates and precisions (Fig. 4a and Supplementary Fig. 7). Our MASH pipeline for modeling dispersion effects was identical to our pipeline for mean effects, with one exception. To identify an initial data-driven set of significant genes (the ‘strong’ set), we selected only genes that passed a q value (FDR-adjusted P value) threshold of 0.05 in five regions (one-third of all tissues) or more (N = 1,940 genes; Supplementary Table 13).
While our focus was on age-related increases in variance of gene expression, we also detected age-associated decreases in variance of gene expression across regions (Fig. 4a and Supplementary Fig. 7). We hypothesized that these results could be attributed in part to global decreases in mean gene expression, which could decrease variance by driving expression to or near zero in older adults. To test this hypothesis, we performed a Fisher’s exact test on a region-wise basis, testing the prediction that genes decreasing in expression with age would be enriched among genes decreasing in dispersion with age (LFSR < 0.2 for both; Supplementary Table 14). Our results supported this prediction, particularly in brain regions for which the majority of aDEGs had negative signs. We therefore proceeded with our main focus on genes increasing in variance with age as putative signatures of age-associated dysregulation.
We tested for enrichment of biological processes and diseases among genes increasing in variance with age. Similar to our analysis of aDEGs, we tested for enrichment of GO biological processes using topGO (Supplementary Fig. 7d and Supplementary Table 15), enrichment of diseases using the DISEASES database (Supplementary Table 16) and enrichment of AD genes using gene lists obtained from the AMP-AD Knowledge Portal24, as detailed earlier (Supplementary Table 17). Because of the relative paucity of significant genes in this analysis after FDR correction, however, we used threshold-independent Kolmogorov–Smirnov (KS) tests to test for enrichment. We ran KS tests using standardized estimates from our MASH model results, calculated by dividing the posterior estimates by the standard deviation of the posterior estimates (standard error). We performed KS tests separately on each brain region and also performed a summary analysis across the brain by averaging standardized beta estimates across all regions in our analysis. For each GO or disease term, we tested the alternative hypothesis that genes associated with each term had significantly more positive standardized beta estimates than other genes. We ran KS tests in topGO using the ‘weight01’ algorithm. For all other analyses, we ran KS tests using custom code in R.
Comparison of brain aging signatures to human bulk RNA-seq data
To validate the aging signatures in our bulk gene expression data and provide a comparison to aging signatures in an orthogonal human dataset, we obtained and analyzed bulk RNA-seq data from the GTEx V8 dataset6. For our analysis, we used as input raw and TPM count matrices from GTEx and retained data from all regions of ‘Brain’ origin with the exception of ‘Brain - Spinal cord (cervical c-1)’. Altogether, we analyzed 2,483 samples from the GTEx dataset, representing 376 individuals (82 females and 233 males; median age = 59.41 years and range = 23–70) and 12 brain regions of similar neuroanatomical scope to our macaque dataset.
Using the same preprocessing pipeline used for our macaque brain data, we transformed raw GTEx counts into log2 (count) values using voom from the limma package83 and removed all lowly expressed genes without an average TPM ≥ 10 in any region, leaving 11,088 total genes.
To compare the relative contributions of age and other biological variables in the human and macaque brain datasets, we performed variance partitioning on the GTEx dataset following nearly identical methods to those described for our macaque data. To control for potentially differing effects of technical variation between datasets, we first removed effects of technical covariates (RNA integrity (SMRIN), genotype/expression batch (SMGEBTCH) and ischemic time (SMTSISCH)) by running linear models on each region separately and extracting partial residuals. Because the GTEx dataset includes two tissue types (‘Brain – Cortex’ and ‘Brain – Cerebellum’) that are dissected separately but are otherwise essentially technical replicates of other regions in the dataset (‘Brain - Frontal Cortex (BA9)’ and ‘Brain - Cerebellar Hemisphere’, respectively), we excluded these two regions to more accurately model the contributions of brain region to overall gene expression. We then ran variance partitioning using the variancePartition package in R, with brain region (SMTSD), age (AGE), sex (SEX) and individual identity (SUB-JID) included as biological variables (Extended Data Fig. 3a). We also ran variance partitioning on each brain region separately to exclude the contribution of region–region variation (Extended Data Fig. 3b).
To compare estimated age effects at the gene level, we modeled gene expression separately for each of the 12 brain regions in our analysis of GTEx data. Because relatedness information was not available, we modeled gene expression and calculated differential expression statistics using lmFit and eBayes from the limma package83. We included age (AGE), sex (SEX), RNA integrity (SMRIN), genotype/expression batch (SMGEBTCH) and ischemic time (SMTSISCH) as covariates. We then used multivariate shrinkage to refine age effect estimates (β), standard errors and LFSRs across the 12 brain regions in mashr, following identical procedures to those described earlier for our macaque brain analysis.
Because we wished to compare GTEx results to previously identified wbaDEGs in our macaque analysis and because our TPM filtering removed a large fraction of these genes, we reran limma and mashr on a small subset of genes (nine macaque wbaDEGs with LFSR < 0.005 in ≥13 regions and having 1:1 orthologs in the human genome) without removing any lowly expressed genes. Using the same covariance matrices estimated from the mashr analysis immediately above, we ran a final model on otherwise-identical gene-by-tissue matrices with this small number of added genes. After examining posterior statistics for the nine genes of interest, we reverted to results from the previous models for downstream analyses.
To determine whether gene expression in rhesus macaque and human brain tissues changes with age in a largely parallel manner, we retained and combined posterior statistics from overlapping brain regions for 8,289 genes that were identified as one-to-one orthologs based on Ensembl annotations104 retrieved using biomaRt94. We compared eight brain–region pairs that overlap between datasets (Supplementary Table 18).
For each brain–region comparison, we compared standardized β estimates (calculated by dividing β by their standard errors) by performing correlation tests using Spearman’s rank correlation coefficient (ρ). We corrected for multiple comparisons by calculating FDR-adjusted P values (q values; Fig. 3 and Supplementary Table 18).
To determine whether signatures of aging parallel one another between macaques and humans at the pathway level, we performed gene set enrichment analysis on wbaDEGs and region-wise aDEGs identified from our analysis of GTEx data and compared the results to those described earlier from our enrichment analysis from macaque data. Apart from using GO annotations for the human (rather than rhesus macaque) genome, we otherwise followed exactly the same procedures described for our macaque analysis.
For 4,013 GO biological process terms that were analyzed in both the macaque and human analyses, we compared enrichment ORs for macaque and human aDEGs identified for all eight overlapping regions and wbaDEGs. We performed comparisons separately for analyses of age-increasing and age-decreasing DEGs (Supplementary Fig. 6 and Supplementary Table 19).
Finally, we aimed to determine whether any components of the macaque and human aging phenotypes either mirrored one another (indicating evolutionary conservatism in aging) or differed from one another (indicating evolutionary divergence in aging) to a greater extent than predicted by chance.
To make this comparison, we estimated sharing within predefined gene sets based on both GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations105. For analysis of GO terms, we used GO annotations as previously described and restricted our analysis to GO biological processes. For analysis of KEGG pathways, we retrieved all KEGG pathways for the rhesus macaque (‘mcc’) genome using KEG-GREST. For both GO and KEGG analyses, we excluded all terms or pathways represented by fewer than 10 genes in our dataset.
For each pathway within each of the eight overlapping brain regions, we characterized sharing as the fraction of genes with shared signs between their estimated age effects between macaques and humans for that region. We estimated significance through permutation. For each permutation iteration, we randomly sampled without replacement an equal number of genes and recalculated the fraction of genes with shared signs from the randomly sampled gene set. We calculated P values as the fraction of null estimates with higher values than the true estimates for each pathway. We performed 10,000 iterations for each pathway/region. We calculated q values using a Benjamini–Hochberg correction95 to correct for multiple hypothesis testing and considered pathways enriched for a given region if they passed a threshold of FDR < 0.05 (Supplementary Tables 20 and 21).
Western blotting
For a subset of animals from our bulk RNA-seq dataset (N = 16), we generated bulk protein lysates from additional frozen 2-mm biopsy punches sampled from the dlPFC (Supplementary Table 22). Biopsy punches were lysed in RIPA buffer (0.05 M Tris-HCl (pH 7.4), 0.5 M NaCl, 0.25% deoxycholic acid, 1% Triton X-100 and 1 mM EDTA; Millipore) supplemented with 0.1% SDS, protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitor cocktail II and III (Sigma-Aldrich). Lysates were cleared by centrifugation, and protein concentration was quantified via the Bio-Rad protein assay (Bio-Rad) using bovine serum albumin as a standard. Equal amounts and volumes of protein were denatured under reducing conditions, separated by SDS–PAGE gels and blotted to PVDF membranes (Bio-Rad). Blots were blocked with 5% low-fat milk in TBS containing 0.1% Tween 20 (TBST) for 1 h at room temperature and incubated overnight at 4 °C with primary antibodies in 4% bovine serum albumin in TBST. The primary antibodies used were anti-FKBP5 (Cell Signaling Technology, 8245; 1:500), anti-MBP (Abcam, ab7349; 1:1,000) and anti-GAPDH (Cell Signaling Technology, 2118S; 1:1,000). After washing with TBST, membranes were incubated with donkey anti-rabbit horseradish peroxidase-conjugated secondary antibodies in 5% milk in TBST for 2 h at room temperature. Blots were washed with TBST, and detection was performed with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). Exposure was manually done in a dark room on radiographic film.
After imaging, film was scanned at high resolution and converted to TIFF format. We then used ImageJ to convert to 8-bit, invert the colors and calculate the integrated density of relevant bands (FKBP5 = ~50 kDa, MBP = ~10–30 kDa and GAPDH = 37 kDa). Equally sized boxes were manually drawn around bands of interest and a blank region of the blot. Each sample lane was background corrected by subtracting the value for the blank region from its respective blot. Blots were mildly stripped and reprobed for the loading control GAPDH. We normalized expression values of MBP and FKBP5 against GAPDH to obtain final values (Supplementary Table 22). Whole blots were optimized for brightness and contrast in Adobe Photoshop (Supplementary Fig. 8).
Immunohistochemistry
We used previously collected formalin-fixed, paraffin-embedded tissue from the dlPFC of two captive macaques (one 7-year-old female and one 22-year-old male) that had been housed at the University of Pennsylvania. Animals were transcardially perfused with 10% formalin, and the brains were immersion fixed for 1–7 d. Sections were dehydrated, embedded in paraffin and sectioned at 4 μm thickness. Before staining, sections were rehydrated and underwent antigen retrieval in a solution containing Tris-EDTA (pH 9.0) that was incubated at 85 °C for 20 min. Sections were then washed with PBS before being blocked in a buffer containing 0.1% Triton X-100 and 5% normal donkey serum in PBS. Next, samples were incubated in a primary antibody solution containing antibodies to NeuN (Millipore, MAB377; 1:1,000) and FKBP5 (Cell Signaling Technology, 8245; 1:300). Sections were then rinsed and incubated in a secondary antibody solution containing anti-mouse Alexa Fluor 568- and anti-rabbit Alexa Fluor 647-conjugated antibodies and DAPI (Sigma-Aldrich, 10236276001; 1:1,000) diluted in blocking solution. Tissue was rinsed in PBS and cover-slipped for microscopic analysis. Images were collected on a Zeiss LSM800 laser-scanning confocal microscope using the same acquisition parameters and were optimized equally for brightness and contrast in Adobe Photoshop (Fig. 2f).
Single-nucleus RNA-seq data generation and quality control
We used sci-RNA-seq3 (ref. 106) to generate single-nucleus 3’ RNA-seq data. The sci-RNA-seq3 protocol uses three rounds of barcoding on intact cells or nuclei to tag transcripts with unique combinatorial indices. The three successives indices are then used to assign transcripts to nuclei, with a low multiplet rate that can be calibrated by controlling the number of nuclei loaded in the experiment and the number of barcodes used in each round of indexing106,107.
Nuclei isolation.
We generated nuclei suspensions from frozen tissue biopsies following methods described by Cao et al.106, with the following modifications. We first homogenized samples by thoroughly grinding samples with a prechilled disposable pestle (Fisher Scientific, 12-141-364) in 50 μl of cell lysis buffer on ice. We brought the volume to 1 ml with ice-cold cell lysis buffer and filtered the mixture using a 70-μm cell strainer (pluriSelect, 43-10070-70). The nuclei were then fixed in 4% paraformaldehyde (6-ml reaction volume) for 15 min with gentle mixing, washed once in 1 ml of ice-cold nuclei wash buffer and resuspended in 200 μl of nuclei wash buffer. After counting a small aliquot with a 1:1 trypan blue stain using the Countess II system, nuclei were allocated in 1-million-nuclei aliquots and flash-frozen in liquid nitrogen.
Library preparation and sequencing.
Methods for sci-RNA-seq3 library preparation and sequencing followed those described by Cao et al.106. Briefly, nuclei were thawed in a 37 °C water bath for 5 min, centrifuged at 500g for 5min and incubated in 400 μl of permeabilization buffer (nuclei wash buffer supplemented with 0.2% Triton X-100) for 3 min. Nuclei were washed, sonicated for 12 s on low power mode (Diagenode), washed and resuspended in 100 μl of nuclei wash buffer. The sci-RNA-seq3 preparation followed the Cao et al. methods for paraformaldehyde-fixed nuclei with the following modifications: samples were counted with the ImageExpress Pico System (Molecular Devices), 2 μl of oligo(dT) primers were added to each well with 80,000 nuclei for reverse transcription, a Quick Ligation kit (New England Biolabs) was used in place of T4 ligase, and tagmentation was performed using 1/40 μl per well of i7-loaded TDE1 enzyme prepared at the Brotman Baty Institute at the University of Washington following published protocols108. Libraries were sequenced on Illumina NovaSeq S4 flow cells at the University of Washington Northwest Genomics Center core sequencing facility with the following cycles: read 1, 34 cycles; read 2, 100 cycles; index 1, 10 cycles; index 2, 10 cycles.
Sequencing data processing.
Single-nucleus RNA-seq reads were processed following the methods described by Cao et al.106 to produce a matrix of unique molecular index (UMI) counts, where rows correspond to genes and columns correspond to unique index combinations, which mainly delineate transcripts from single nuclei but also include a small fraction of multiplets. Our pipeline was modified for the most recent rhesus macaque reference genome (Mmul_10)81 and annotation obtained from Ensembl (version 101). Haplotype sequences were removed from the genome FASTA to avoid mapping inefficiencies due to multimapping, and the 3′-untranslated region annotations of genes and transcripts were extended in the reference transcriptome (GTF file) by 500 bp to avoid misclassifying genic reads as intergenic. We proceeded with the remainder of the Cao et al. sci-RNA-seq3 pipeline for demultiplexing, mapping, deduplication and count matrix generation using the 3′-untranslated region extended GTF file as input. We used monocle 3 (ref. 106) to generate a cell dataset (cds) object.
Genotyping.
Similar to our bulk RNA-seq analyses, we inferred genotypes from sci-RNA-seq3 reads to estimate relatedness among animals. We combined all non-redundant reads (reads with unique combinatorial indices and UMIs) per animal after first trimming adapter and barcode sequences from the reads. We then mapped reads to the rhesus macaque reference genome using STAR86. Because high duplication rates and the lower abundance of nuclear transcripts limit read depths relative to bulk RNA-seq, we inferred genotypes using ngsRelate2, a method incorporating genotype uncertainty that performs well even with low sequencing depth109,110. We used ANGSD111 to estimate allele frequencies and generate genotype likelihoods, estimating major and minor alleles from the data and using a minimum minor allele frequency (minmaf) of 0.05 and a P value threshold (SNP_pval) of 1 × 10−6. We then ran ngsRelate2 using the resulting allele frequencies and genotype likelihoods. We used resulting estimates of pairwise relatedness (rab)112 to build a relatedness matrix.
Quality control and filtering.
We performed an initial filtering of our gene χ cell UMI matrix by excluding from our analysis nuclei with fewer than 100 UMIs or greater than 10% of reads mapping to the mitochondrial genome (Supplementary Fig. 9a-c). We then used Scrublet113 to identify multiplets on a per-sample basis. For each sample, we visualized the distribution of k-nearest neighbor (kNN) scores as a density plot and manually set thresholds to remove likely multiplets (Supplementary Fig. 9d). To validate multiplet removal, we performed preprocessing, UMAP dimensionality reduction and Leiden clustering in monocle 3 (ref. 106) to ensure clean separation and clustering of distinct types. Additionally, we performed preprocessing, UMAP dimensionality reduction and Leiden clustering on the entire dataset, assessed kNN score distributions across clusters and manually removed clusters with noticeably high kNN score distributions (Supplementary Fig. 9e,f). After removing all likely multiplets, we performed a final UMAP dimensionality reduction and Leiden clustering (k = 20) on the entire dataset containing 71,863 unique indices (Fig. 5a and Supplementary Fig. 10), which we refer to henceforth as nuclei.
Cell type and cell subtype identification.
We assigned nuclei to one of eight broad brain cell types using previously described marker genes114 and from top marker analysis of published single-nucleus RNA-seq datasets115 (Supplementary Table 23). We manually assigned cell types to each cluster by visualizing the specificity and breadth of expression of each marker gene. To validate our cell-type assignments, we used the data integration workflow in Seurat v3 (ref. 116) to harmonize our data with a subset of 200,000 randomly sampled cells from the Allen Brain Map Mouse Whole Cortex and Hippocampus 10x dataset117-119 as well as 76,533 cells from the Allen Brain Map human M1 10x dataset42. We then used SingleR120 to perform label transfer on the integrated data space. The integrated labels largely corroborated our cell-type assignments (Supplementary Fig. 11 and Supplementary Tables 24 and 25).
To identify cell subtypes, we subset the gene × cell UMI matrix for each cell type separately and reperformed UMAP dimensionality reduction and Leiden clustering (k = 5) in monocle 3. We designated each resulting cluster as a distinct cell subtype (Supplementary Fig. 10a).
For both cell types and cell subtypes, we determined marker genes using top_markers from monocle 3 (ref. 106). We summarized each cell type or subtype by selecting the top 20 markers based on the pseudo-R2 statistic121 (Supplementary Tables 26 and 27).
Estimating age effects on cell-type proportions.
For each individual, we calculated cell proportions as the number of nuclei assigned to each cell type or subtype divided by the total number of nuclei for that individual. We then modeled cell proportions using linear models with age and social group as covariates (Fig. 5b, Extended Data Fig. 6b and Supplementary Tables 28 and 29). We calculated Bonferroni-adjusted P values to correct for multiple hypothesis testing.
Cell-type deconvolution of bulk gene expression
To distinguish between age-associated changes in mean expression within cells and age-associated changes in cell-type composition of our bulk RNA-seq dataset, we used our single-nucleus RNA-seq dataset as a reference dataset for cell-type deconvolution via the R package BRETIGEA122. To determine cell-type marker genes, we used top_markers from monocle 3 (ref. 106), selecting the top 50 markers for each cell cluster (cell subtype) based on the pseudo-R2 statistic121. Finally, for each brain region, we performed principal components analysis on resulting estimates of bulk RNA-seq library cell-type proportions (derived by summing proportion estimates across all subtypes), retaining the first N principal components that together explained greater than 90% of the variance. We included these principal components as covariates and repeated our EMMA and MASH pipelines to estimate age effects from our deconvoluted bulk RNA-seq data on a per-brain-region basis. We found that age effects were highly correlated between the analyses with and without cell-type deconvolution, with slightly weaker effect sizes with cell-type deconvolution reflecting some age-associated changes in cell-type proportion (Extended Data Fig. 8b).
Estimating linear age effects within cell types
To model age-associated differences in gene expression among cell types, we bioinformatically converted our single-cell dlPFC data into pseudobulk libraries by summing gene-wise UMI counts within seven broad cell classes: excitatory neurons, inhibitory neurons, astrocytes, oligodendrocytes, oligodendrocyte precursor cells, microglia and endothelial cells. We excluded pericytes from subsequent analyses because only 266 nuclei were assigned to this cell type, with two individuals lacking any assigned pericytes.
Similar to our bulk tissue RNA-seq analysis, for each cell type, we excluded all genes having a mean counts per million of <10 in that cell type and calculated log2 (count) values using voom from the limma package83. We observed, however, that cells experiencing age-related changes in cell proportion also exhibited global differences in normalized gene expression (Supplementary Fig. 12), which is likely an artifact due to higher degrees of zero inflation in lower-coverage pseudobulk libraries generated from less-abundant cell types. We thus instead normalized our pseudobulk expression data with geometric mean of pairwise ratios (GMPR), a normalization method developed for zero-inflated microbiome sequencing count data123. Unlike other techniques for mitigating zero inflation artifacts, GMPR does not assume that zeroes are due to undersampling and is flexible for datasets with high count variability123. These qualities are particularly salient for pseudobulk analyses of single cell types, where zeroes may also be explained by true lack of expression, and high count variability can be driven by differential cell abundance and by real biological processes. After normalizing our data with GMPR, we confirmed the absence of spurious relationships between cell proportion and global gene expression changes (Supplementary Fig. 12).
We modeled GMPR-normalized log2 (count) values using EMMA models10,96 implemented in R using EMMREML (Extended Data Fig. 7a). For all models, we included age, social group and extraction batch as fixed effects and used the custom kinship matrix generated for this dataset described previously as random effects. We then used MASH to pool information across cell types and to refine our estimates of effect size and precision (Fig. 5d, Extended Data Fig. 7b and Supplementary Table 31).
We tested for enrichment of GO biological processes (Supplementary Table 32) among effects estimates for single cell types using topGO92,93. In contrast to our bulk tissue analysis, however, we tested for significance using threshold-independent KS tests because the smaller sample size of this analysis resulted in fewer genes passing thresholds. We ran KS tests in topGO using the weight01 algorithm.
To replicate our bulk tissue RNA-seq results, we summed gene-wise UMI counts for each individual (that is, combining across all cell types) to generate pseudobulk libraries at the tissue level, which we processed and modeled using the same pipeline described for each cell type separately. We then plotted corresponding model estimates for all genes from our true bulk and pseudobulk dlPFC analyses (Extended Data Fig. 9c).
Estimation of shared effects between social adversity and aging
To test for shared effects of aging and social adversity, we compared effect sizes of age and dominance rank on regional gene expression. We estimated effect sizes of dominance rank using MASH11 following the procedures described earlier to pool information across tissues and refine effect size estimates and LFSRs for the dominance rank term from our EMMA model.
Because we predict parallel changes with older age and higher social adversity (that is, lower dominance rank), we reversed the signs for the ordinal dominance rank effect sizes before comparison so that genes with higher expression in low-ranking animals had positive values. Positive effect sizes from our models thus indicate increased expression with older age and lower rank, respectively.
For each region, we estimated the degree of sharing between aging and social adversity by calculating Spearman’s correlation (ρ). Because determining the degree of sharing requires a tradeoff between the number ofgenes included and the statistical precision of the effect size estimates, we repeated this analysis across a range of LFSR thresholds (θ) from 0 to 1 in 0.01 increments for which we only included genes with an LFSR of ≤θ for both age and dominance rank (Fig. 6a and Supplementary Fig. 13). For each estimate, we used 1,000 bootstrap iterations to calculate 95% confidence intervals (Supplementary Fig. 13b).
To characterize shared effects between aging and social adversity at the pathway level, we followed an approach similar to our comparison of macaque (this study) and human (GTEx study) age effects but modified to compare aging and dominance rank effects within the macaque dataset. We estimated sharing within predefined gene sets based on both GO and KEGG annotations and filtered as described previously for our comparison to GTEx data.
For each pathway within each brain region, we characterized sharing as the fraction of genes with shared signs between their estimated age and dominance rank effects for that region. We then calculated P values and FDR-adjusted P values (q values) through permutation. We considered pathways enriched for a given region if they passed a threshold of FDR < 0.05 (Supplementary Tables 33 and 34). Notably, our permutation-based estimation of significance is conservative due to background positive correlations between aging and dominance rank effects across most of our dataset (Fig. 6a and Supplementary Fig. 13).
Dominance rank effects on transcriptomic age
We tested the prediction that experiencing social adversity (that is, having low dominance rank) accelerates aging, resulting in an older transcriptomic age phenotype than predicted by chronological age. To test this prediction, we used two approaches to predict transcriptomic age, which we refer to here as our ‘wbaDEGs’ and ‘glmnet’ approaches.
For the first (wbaDEGs) approach, we used our age coefficients estimated by EMMA models and refined by MASH. To develop a universal transcriptomic age predictor across all brain regions, we kept only genes that passed an LFSR threshold and whose effect estimates shared signs in at least 13 of 15 regions. After selecting the most universally age-predictive genes, we constructed age predictors by averaging effect estimates across all 15 regions for each gene and calculated transcriptomic ages from our z-scored gene expression matrix using the equation given by Peters et al.124, previously used by our group on peripheral blood data2. We repeated this procedure across a range of LFSR thresholds and selected the threshold at which average mean absolute error was minimal (LFSR < 0.005; Supplementary Fig. 14b). We thus obtained a set of 13 genes fulfilling these criteria. We then used leave-one-out cross-validation (LOOCV) to ensure independence of our training and evaluation data. For each LOOCV iteration, we excluded all samples from one animal from our analysis and reran our EMMA and MASH models following procedures described above. We then recalculated our 13 age predictors as described above and calculated unscaled transcriptomic age predictions from the z-scored expression matrix of all samples from the animal withheld from model training. We converted unscaled age predictions from all samples estimated in this manner into transcriptomic age predictions by scaling values to the known age distribution in our population124 (Fig. 6b and Supplementary Fig. 14a).
As an additional proof of concept, we used our wbaDEGs age prediction coefficients from 13 genes to predict ages from our pseudobulk bulk tissue dlPFC libraries (Extended Data Fig. 9d).
For the second (glmnet) approach, we used an elastic net regression model to predict ages from our z-scored gene expression matrices containing libraries from all regions. We fit elastic net models using the glmnet package125 in R using a Gaussian error distribution and α = 0.5 to evenly balance ridge and lasso regularization. We ran an initial elastic net model with 10-fold cross-validation to estimate the λ parameter. For each region, we ran elastic net models with an animal-wise version of LOOCV to predict ages. For each iteration, we withheld all libraries from an animal as our test sample and trained our model from all libraries from the remaining animals. We predicted ages on our test samples using the λ values estimated from the initial model (Supplementary Fig. 14).
For both approaches, we originally estimated aging phenotypes for each individual as the difference between its predicted transcriptomic age and its known chronological age (absolute error). We refer to this difference as ΔAge. Because we found that for both approaches, the range of transcriptomic age predictions was narrower than the chronological age range of our sample, our ΔAge estimates had strong non-zero relationships with chronological age. We therefore instead estimated aging phenotypes by first fitting a linear least-squares regression and using the residuals of the model as an alternative to ΔAge (ref. 126). The resulting measure eliminates any confounding relationship with chronological age. We call this measure eAge.
We tested the prediction that social adversity accelerates aging by regressing eAge for each sample against percent dominance rank and categorical dominance rank, separately, using linear mixed models controlling for individual-level random variation and the fixed effect of brain region. We adjusted P values for one-sided hypothesis testing using the t distribution for all tests.
Chosen genes that are identified as extremely predictive for either age prediction approach are not necessarily linked to putative dominance rank effects on aging and may represent a relatively miniscule component of the overall aging landscape. We reasoned that genes demonstrating strong relationships with both dominance rank and chronological age are more likely to be functionally linked to possible dominance rank effects on aging. We developed an approach in which we iteratively implemented increasingly strict dominance rank-based filters to limit an initial gene list and reran both our wbaDEGs and glmnet age prediction pipelines (including LOOCV) to predict ages only from these initial gene lists with increasingly well-supported dominance rank effects. This approach necessarily requires a tradeoff between our confidence/precision in dominance rank effect estimates and the age-predictive performance of the models, which by definition decreases with increasingly strict thresholds because greater numbers of age-predictive genes that would otherwise be selected are withheld from the model.
For our wbaDEGs models, we imposed filters within the range LFSRRank < (0.01, 0.001, 0.0001). For genes passing each filter, we averaged age effect estimates across all 15 brain regions and used these coefficients to predict age as described earlier. For our glmnet models, we imposed filters within the range LFSRRank < (0.2, 0.1, 0.05) and reran glmnet model training and prediction as described earlier. For both approaches, we retained genes if they passed a given threshold in any brain region (Fig. 6c,d, Supplementary Fig. 15 and Supplementary Tables 36 and 37).
Given that increasingly strict filtering comes with a cost to predictive performance, it is plausible that filtering could eventually exhaust any biological signal, resulting in noisy age predictions with unreliable residuals. To rule out this possibility while simultaneously ensuring that observed patterns were robust to thresholding, we repeated our analyses across a greater range of LFSRRank thresholds. For our wbaDEG models, we tested all thresholds 10−i, where i ranged from 0 to 8 in 0.5 increments. For our glmnet models, we tested all thresholds 10−i, where i ranged from 0 to 2 in 0.1 increments. For each iteration, after predicting ages as described above, we performed (1) a Pearson’s correlation test assessing the strength of correlation between chronological and predicted ages and (2) a linear mixed model assessing the relationship between percent dominance rank and residual age, as described earlier. We then jointly visualized results from these two tests to confirm that there is sufficient biological signal across a range of thresholds (Supplementary Fig. 16).
Extended Data
Extended Data Fig. 1 ∣. Sample and study design.
(a) Location of Cayo Santiago off the southeastern coast of Puerto Rico. (b) Age distribution of individuals sampled for the bulk tissue RNA sequencing dataset. Selected individuals span the natural adult age distribution and are evenly balanced between sexes. (c) Age distribution of individuals per brain region (excludes samples not passing laboratory or bioinformatic quality control). (d) Age distribution of females sampled for the single-nucleus RNA sequencing dataset. Selected individuals span the natural age distribution and are evenly balanced between two naturally occurring social groups (HH and KK) on Cayo Santiago. (e) Hazard rates from a previously published demographic model of Cayo Santiago macaques (ref. 2) demonstrate that individuals in the age range of our sample experience age-associated increases in mortality risk (N = 11,659 biologically independent animals). Error bars represent the mean hazard rate ± the standard error.
Extended Data Fig. 2 ∣. Global similarity of gene expression across brain regions.
(a) Dendrogram showing hierarchical clustering results on averaged expression for each brain region. (b) The same dendrogram visualized over 1000 bootstrap replicates in which libraries were randomly sampled with replacement before calculating average expression and repeating hierarchical clustering. (c) Dendrogram showing hierarchical clustering results on all libraries. Terminal branches are colored by brain region.
Extended Data Fig. 3 ∣. Comparison of variance partitioning of age between macaques and humans.
(a) Age explains similar proportions of variance in global gene expression across diverse brain regions between macaques from this study (left) and humans from the GTEx study (right). Median variance explained by age was 0.19% (interquartile range [IQR] = 0.04–0.51%) in this study (N = 527 biologically independent samples) and 0.26% (IQR = 0.07–0.82%) in the GTEx study (N = 2,642 biologically independent samples). Effects of technical covariates were first removed to facilitate this comparison. (b) Age explains similar proportions of variance in gene expression within individual brain regions. Median variance explained by age ranged from 0.6–6.4% across brain regions from this study (N = 36 biologically independent animals, left) and ranged from 0.3–4.2% across brain regions from the GTEx study (N = 382 biologically independent individuals, right). Box plots depict the median (center), and IQR (bounds of box), with whiskers extending to either the maxima/ minima or to the median ±1.5×IQR, whichever is nearest.
Extended Data Fig. 4 ∣. Analysis of age-associated effects on gene expression levels.
(a) Quantile-quantile (QQ) plots of P values from initial efficient mixed model association (EMMA) tests show enrichment of low P values across nearly all brain regions tested (all EMMA tests are two-sided). (b) A multivariate adaptive shrinkage (MASH) approach substantially improves statistical power by leveraging shared patterns between tissue datasets, resulting in a greater number of genes passing a threshold (local false sign rate [LFSR] < 0.2) relative to a similar threshold applied to our EMMA results (false discovery rate [FDR] < 0.2). (c) The number of significant genes visualized over a range of LFSR thresholds shows extremely stable rank order of brain regions. (d) Whole-brain age-differentially expressed genes (wbaDEGs) cross-referenced with our gene trajectory results (Fig. 1d) demonstrate that the vast majority of wbaDEGs fall into four clusters, marked by asterisks (***). Most wbaDEGs decreasing in expression with age fall into trajectories associated with signaling-related functions, while most wbaDEGs increasing in expression with age fall into a trajectory associated with the immune response. In the bottom panel, the percentage of genes assigned to wbaDEGs is plotted on the y axis.
Extended Data Fig. 5 ∣. Similarities of age effects across brain regions.
(a) Proportion of aDEGs with shared magnitude between brain regions. aDEGs share magnitude between regions when they share signs and their effect estimates are within a factor of 2 from one another. These conditions for sharing are more stringent than those in Fig. 2b, which does not require the latter criterion. For each pair of regions, genes are included if they were significant (LFSR < 0.2) in either region. (b) Upset plot highlights brain region combinations with the greatest number of aDEGs with shared signs. Note that among the top region combinations are single-region-specific aDEGs as well as wbaDEGs. (c) Proportion of aDEGs that exhibit (top) shared signs or (bottom) shared magnitudes across variable numbers of brain regions. These distributions reveal that subcortical regions sampled exhibited slightly broader sharing than cortical regions. For this analysis, genes are included if they were significant (LFSR < 0.2) in any region.
Extended Data Fig. 6 ∣. Cell-type marker genes and proportional changes with age.
(a) Expression of top 5 marker genes (according to the pseudo-R2 statistic) for 8 assigned cell types. (b) Age plotted against cell cluster percentages. Asterisks in cluster titles indicate a significant effect of age on cell cluster percentages based on a linear model (two-sided test, excitatory neurons 6: Bonferroni-adjusted P = 0.014; excitatory neurons 10: Bonferroni-adjusted P = 0.024).
Extended Data Fig. 7 ∣. Age-associated differences in gene expression within single cell types.
(a) QQ plot of EMMA tests on pseudobulk data show a strong enrichment of low P values (all EMMA tests are two-sided). (b) Comparison of model estimates from EMMA (FDR < 0.2) and MASH (LFSR < 0.2) show similar numbers of significant genes. (c). Cell-type aDEG counts (MASH) classified according to overlap with expressed genes and aDEGs from bulk RNA-seq analysis of the dlPFC. (d). Percentages of aDEGs overlapping with aDEGs from bulk RNA-seq analysis of the dlPFC (excluding genes not expressed in both modalities). (e) Rank order of significant genes per cell type are robust across a range of significance (LFSR) thresholds. (f) Upset plot shows rank order of cell type combinations with the greatest number of genes exhibiting significant age-associated differences in expression.
Extended Data Fig. 8 ∣. Cell-type deconvolution of bulk tissue RNA-seq data.
(a) After adjusting for cell type composition and repeating our EMMA and MASH models, similar numbers of genes per brain region pass our significance thresholds (LFSR < 0.2). (b) Standardized age effect estimates from analyses controlling and not controlling for cell-type composition are strongly positively correlated, but slopes < 1 in most brain regions suggest that some age effects identified in our bulk gene expression results are due to age-related changes in cell proportion. (c) The majority of aDEGs belonging to four clusters with pronounced age-associated directional changes (Fig. 1d) met our criteria (LFSR < 0.2) as aDEGs after controlling for heterogeneous tissue compositions using cell-type deconvolution (CD) analysis. Substantial fractions, however, did not, providing further evidence that some results at the bulk-tissue level are driven by age-related changes in cell proportion.
Extended Data Fig. 9 ∣. Exploratory analyses of single-cell-type pseudobulk data from the dlPFC.
UMAP plots of pseudobulk libraries reveal that the latent structure is driven primarily by (a) cell type and not (b) age. (c) Single-nucleus libraries were aggregated across all cell types to approximate bulk tissue RNA sequencing libraries. Age effects from pseudobulk dlPFC libraries were then compared to age effects from bulk dlPFC libraries, showing a strong positive correlation. Colors denote whether age effects were considered significant for both datasets (EMMA test uncorrected P < 0.05). Error bands represent the 95% confidence interval of linear model predictions. (d) Similarly, age prediction on pseudobulk libraries using the wbaDEGs model (see Methods) developed from analysis of bulk RNA-seq data shows a similar positive correlation (linear model statistics are presented) and accuracy to predictions on bulk tissue libraries. Error bands represent the 95% confidence interval of linear model predictions.
Supplementary Material
Acknowledgements
We thank those who make our research possible, particularly the CPRC and the Cayo Santiago Field Station staff, without whom we would not be able to study this amazing system. We also thank the staff of the Sabana Seca Field Station for assistance with sample collection and J. Cao, A. Lea, N. Simons, R. Campbell, J. Tung, I. Schneider-Crease and the University of Washington Basic Biology of Aging training group for valuable feedback at various stages throughout the project. Funding for this research was provided by the National Institutes of Health (R01-MH118203 to M.L.P., U01-MH121260 to N.S.-M., M.L.P., and J. Shendure, R01-MH096875 to M.L.P., R01-AG060931 to N.S.-M., L.J.N.B., and J.P.H., R00-AG051764 to N.S.-M., R01-NS097537 to J.M.N., R35-GM124827 to M.A.W., K99-AG075241 to K.L.C. and P40-OD012217 to M.I.M.), the National Science Foundation (BCS-1800558 to J.P.H. and BCS-1752393 to A.R.D.) and a pilot grant to N.S.-M. from the Brotman Baty Institute. K.L.C. was supported by National Institutes of Health fellowship T32-AG000057 during this research.
The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (https://commonfund.nih.gov/GTEx). Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI\Leidos Biomedical Research, Inc., subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171) and Science Care, Inc. (X10S172). The Laboratory, Data Analysis and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through a Leidos Biomedical Research, Inc., subcontract to Van Andel Research Institute (10ST1035). Additional data repository and project management were provided by Leidos Biomedical Research. (HHSN261200800001E). The Brain Bank was supported by supplements to University of Miami grant DA006227. Statistical Methods development grants were made to the University of Geneva (MH090941 and MH101814), the University of Chicago (MH090951, MH090937, MH101825 and MH101820), the University of North Carolina Chapel Hill (MH090936), North Carolina State University (MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University (MH101810) and the University of Pennsylvania (MH101822). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000424.v8.p2.
The results published here are in whole or in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.org/). MayoRNAseq data were provided by the following sources: The Mayo Clinic Alzheimer’s Disease Genetic Studies, led by N. Taner and S. G. Younkin, Mayo Clinic, Jacksonville, FL, using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimer’s Disease Research Center and the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216 and R01 AG003949, NINDS grant R01 NS080820, CurePSP Foundation and support from Mayo Foundation. Study data include samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, AZ. The Brain and Body Donation Program is supported by the NINDS (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the NIA (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. MSBB data were generated from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by E. Schadt from Mount Sinai School of Medicine. ROSMAP data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, IL. Data collection was supported through funding by NIA grants P30AG10161 (ROS), R01AG15819 (ROSMAP; genomics and RNA-seq), R01AG17917 (MAP), R01AG30146, R01AG36042 (5hC methylation and ATACseq), RC2AG036547 (H3K9Ac), R01AG36836 (RNA-seq), R01AG48015 (monocyte RNA-seq) RF1AG57473 (single-nucleus RNA-seq), U01AG32984 (genomics and whole-exome sequencing), U01AG46152 (ROSMAP AMP-AD and targeted proteomics), U01AG46161 (TMT proteomics), U01AG61356 (whole-genome sequencing, targeted proteomics and ROSMAP AMP-AD), the Illinois Department of Public Health (ROSMAP) and the Translational Genomics Research Institute (genomics). Additional phenotypic data can be requested at www.radc.rush.edu.
Cayo Biobank Research Unit
Susan C. Antón5,6, Lauren J. N. Brent22, James P. Higham5,6, Melween I. Martínez11, Amanda D. Melin14,15,16, Michael J. Montague7, Michael L. Platt7,23,24, Jérôme Sallet18 & Noah Snyder-Mackler1,2,3,4,25,26,27
Footnotes
Competing interests
J. Shendure is a scientific advisory board member, consultant and/or cofounder of Cajal Neuroscience, Guardant Health, Maze Therapeutics, Camp4 Therapeutics, Phase Genomics, Adaptive Biotechnologies, Scale Biosciences and Sixth Street Capital. M.L.P. is a scientific advisory board member, consultant and/or cofounder of Blue Horizons International, NeuroFlow, Amplio, Cogwear Technologies, Burgeon Labs and Ashurst Cognitive Health and receives research funding from AIIR Consulting, the SEB Group, Mars Inc., Slalom Inc., the Lefkort Family Research Foundation, Sisu Capital and Benjamin Franklin Technology Partners. All other authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41593-022-01197-0.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41593-022-01197-0.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41593-022-0n97-0.
A list of authors and their affiliations appears at the end of the paper.
Data availability
The data generated in this study can be downloaded in raw and processed forms from the NCBI Gene Expression Omnibus under series accession number GSE179330.
Datasets/databases used in this study are publicly available and include the rhesus macaque Mmul_10 reference assembly (NCBI RefSeq accession GCF_003339765.1), Ensembl release 101 (http://aug2020.archive.ensembl.org.), KEGG release 100.0 (https://www.genome.jp/kegg), GTEx analysis V8 (dbGaP accession phs000424.v8.p2) and the Alzheimer’s Knowledge Portal AMP-AD meta-analysis (https://doi.org/10.7303/syn11914606).
All requests for biological material should be directed to the CBRU (cbru@pennmedicine.upenn.edu). Requests will be reviewed by the Scientific Review Committee and granted depending on availability, proposed use, investigator funding and other considerations.
Code availability
All code for this study is accessible through the following GitHub repositories: https://github.com/CayoBiobankResearchUnit/brain_transcriptome_aging_bulk (bulk RNA-seq analysis), https://github.com/CayoBiobankResearchUnit/brain_transcriptome_aging_sc (single-nucleus RNA-seq analysis), https://github.com/bbi-lab/bbi-dmux (sci-RNA-seq3 data demultiplexing) and https://github.com/bbi-lab/bbi-sci. (sci-RNA-seq3 data preprocessing).
References
- 1.Hou Y et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol 15, 565–581 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Chiou KL et al. Rhesus macaques as a tractable physiological model of human ageing. Philos. Trans. R. Soc. Lond. B Biol. Sci 375, 20190612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernandez-Pacheco R et al. Managing the Cayo Santiago rhesus macaque population: the role of density. Am. J. Primatol 78, 167–181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider JA, Arvanitakis Z, Leurgans SE & Bennett DA The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann. Neurol 66, 200–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair JA et al. Individual case analysis of postmortem interval time on brain tissue preservation. PLoS ONE 11, e0151615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaum N et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature 583, 596–602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deleidi M, Jäggle M & Rubino G Immune aging, dysmetabolism, and inflammation in neurological diseases. Front. Neurosci 9, 172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mecocci P et al. A long journey into aging, brain aging, and Alzheimer’s disease following the oxidative stress tracks. J. Alzheimers Dis 62, 1319–1335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sul JH, Martin LS & Eskin E Population structure in genetic studies: confounding factors and mixed models. PLoS Genet. 14, e1007309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urbut SM, Wang G, Carbonetto P & Stephens M Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat. Genet 51, 187–195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens M False discovery rates: a new deal. Biostatistics 18, 275–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheff SW, Price DA, Schmitt FA & Mufson EJ Hippocampal synaptic loss in early Alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging 27, 1372–1384 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Poulin SP et al. Amygdala atrophy is prominent in early Alzheimer’s disease and relates to symptom severity. Psychiatry Res. 194, 7–13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder EB et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat. Genet 36, 1319–1325 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Sinclair D, Fillman SG, Webster MJ & Weickert CS Dysregulation of glucocorticoid receptor co-factors FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness. Sci. Rep 3, 3539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blair LJ et al. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Invest 123, 4158–4169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halbgebauer S et al. Modified serpinA1 as risk marker for Parkinson’s disease dementia: analysis of baseline data. Sci. Rep 6, 26145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebbert MTW et al. Conserved DNA methylation combined with differential frontal cortex and cerebellar expression distinguishes C9orf72-associated and sporadic ALS, and implicates SERPINA1 in disease. Acta Neuropathol. 134, 715–728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chai Z, Zheng P & Zheng J Mechanism of ARPP21 antagonistic intron miR-128 on neurological function repair after stroke. Ann. Clin. Transl. Neurol 8, 1408–1421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Cooper-Knock J et al. Mutations in the glycosyltransferase domain of GLT8D1 are associated with familial amyotrophic lateral sclerosis. Cell Rep. 26, 2298–2306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwanhäusser B et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Beyer A & Aebersold R On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Greenwood AK et al. The AD Knowledge Portal: a repository for multi-omic data on Alzheimer’s disease and aging. Curr. Protoc. Hum. Genet 108, e105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izgi H et al. Inter-tissue convergence of gene expression during ageing suggests age-related loss of tissue and cellular identity. eLife 11, e68048 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland PWH & Takahashi T The evolution of homeobox genes: implications for the study of brain development. Brain Res. Bull 66, 484–490 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Bergsland M et al. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 25, 2453–2464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould E How widespread is adult neurogenesis in mammals? Nat. Rev. Neurosci 8, 481–488 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Diederich NJ, James Surmeier D, Uchihara T, Grillner S & Goetz CG Parkinson’s disease: is it a consequence of human brain evolution? Mov. Disord 34, 453–459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pouladi MA, Morton AJ & Hayden MR Choosing an animal model for the study of Huntington’s disease. Nat. Rev. Neurosci 14, 708–721 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Finch CE & Austad SN Commentary: is Alzheimer’s disease uniquely human? Neurobiol. Aging 36, 553–555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yerbury JJ et al. Walking the tightrope: proteostasis and neurodegenerative disease. J. Neurochem 137, 489–505 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Nativio R et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci 21, 497–505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinney JW et al. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement. 4, 575–590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vicario-Orri E, Opazo CM & Muñoz FJ The pathophysiology of axonal transport in Alzheimer’s disease. J. Alzheimers Dis 43, 1097–1113 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Eschbach J & Dupuis L Cytoplasmic dynein in neurodegeneration. Pharmacol. Ther 130, 348–363 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Glass CK, Saijo K, Winner B, Marchetto MC & Gage FH Mechanisms underlying inflammation in neurodegeneration. Cell 140, 918–934 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan Y-W et al. Meta-analysis of the Alzheimer’s disease human brain transcriptome and functional dissection in mouse models. Cell Rep. 32, 107908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S et al. Extent of dorsolateral prefrontal cortex plasticity and its association with working memory in patients with Alzheimer disease. JAMA Psychiatry 74, 1266–1274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Upright NA & Baxter MG Prefrontal cortex and cognitive aging in macaque monkeys. Am. J. Primatol 83, e23250 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Prater KE et al. Subtype transcriptomic profiling of myeloid cells in Alzheimer disease brain illustrates the diversity in active microglia phenotypes. Preprint at bioRxiv 10.1101/2021.10.25.465802 (2021). [DOI] [Google Scholar]
- 42.Bakken TE et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luebke J, Barbas H & Peters A Effects of normal aging on prefrontal area 46 in the rhesus monkey. Brain Res. Rev 62, 212–232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raible DW & McMorris FA Induction of oligodendrocyte differentiation by activators of adenylate cyclase. J. Neurosci. Res 27, 43–46 (1990). [DOI] [PubMed] [Google Scholar]
- 45.Perry VH & Holmes C Microglial priming in neurodegenerative disease. Nat. Rev. Neurol 10, 217–224 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Niraula A, Sheridan JF & Godbout JP Microglia priming with aging and stress. Neuropsychopharmacology 42, 318–333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowsky DJ, Olshansky SJ, Bhattacharya J & Goldman DP Heterogeneity in healthy aging. J. Gerontol. A Biol. Sci. Med. Sci 69, 640–649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belsky DW et al. Quantification of biological aging in young adults. Proc. Natl Acad. Sci. USA 112, E4104–E4110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snyder-Mackler N et al. Social determinants of health and survival in humans and other animals. Science 368, eaax9553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blomquist GE, Sade DS & Berard JD Rank-related fitness differences and their demographic pathways in semi-free-ranging rhesus macaques (Macaca mulatta). Int. J. Primatol 32, 193–208 (2011). [Google Scholar]
- 51.Snyder-Mackler N, Somel M & Tung J Shared signatures of social stress and aging in peripheral blood mononuclear cell gene expression profiles. Aging Cell 13, 954–957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Testard C et al. Rhesus macaques build new social connections after a natural disaster. Curr. Biol 31, 2299–2309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McColgan P, Joubert J, Tabrizi SJ & Rees G The human motor cortex microcircuit: insights for neurodegenerative disease. Nat. Rev. Neurosci 21, 401–415 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Ohm TG The dentate gyrus in Alzheimer’s disease. Prog. Brain Res 163, 723–740 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Jiji S, Smitha KA, Gupta AK, Pillai VPM & Jayasree RS Segmentation and volumetric analysis of the caudate nucleus in Alzheimer’s disease. Eur. J. Radiol 82, 1525–1530 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Wilson RS et al. Loneliness and risk of Alzheimer disease. Arch. Gen. Psychiatry 64, 234–240 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Holwerda TJ et al. Feelings of loneliness, but not social isolation, predict dementia onset: results from the Amsterdam Study of the Elderly (AMSTEL). J. Neurol. Neurosurg. Psychiatry 85, 135–142 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Cadar D et al. Individual and area-based socioeconomic factors associated with dementia incidence in England: evidence from a 12-year follow-up in the English Longitudinal Study of Ageing. JAMA Psychiatry 75, 723–732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berard J A four-year study of the association between male dominance rank, residency status, and reproductive activity in rhesus macaques (Macaca mulatta). Primates 40, 159–175 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Zannas AS et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 16, 266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zannas AS Epigenetics as a key link between psychosocial stress and aging: concepts, evidence, mechanisms. Dialogues Clin. Neurosci 21, 389–396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harvanek ZM, Fogelman N, Xu K & Sinha R Psychological and biological resilience modulates the effects of stress on epigenetic aging. Transl. Psychiatry 11, 601 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller GE, Cohen S & Ritchey AK Chronic psychological stress and the regulation of pro-inflammatory cytokines: a glucocorticoid-resistance model. Health Psychol. 21, 531–541 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Snyder-Mackler N et al. Social status alters immune regulation and response to infection in macaques. Science 354, 1041–1045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snyder-Mackler N et al. Social status alters chromatin accessibility and the gene regulatory response to glucocorticoid stimulation in rhesus macaques. Proc. Natl Acad. Sci. USA 116, 1219–1228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kessler MJ & Rawlins RG A 75-year pictorial history of the Cayo Santiago rhesus monkey colony. Am. J. Primatol 78, 6–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Missakian EA Genealogical and cross-genealogical dominance relations in a group of free-ranging rhesus monkeys (Macaca mulatta) on Cayo Santiago. Primates 13, 169–180 (1972). [Google Scholar]
- 68.Widdig A et al. Low incidence of inbreeding in a long-lived primate population isolated for 75 years. Behav. Ecol. Sociobiol 71, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finch CE & Austad SN Primate aging in the mammalian scheme: the puzzle of extreme variation in brain aging. Age 34, 1075–1091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roth GS et al. Aging in rhesus monkeys: relevance to human health interventions. Science 305, 1423–1426 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Kessler MJ, Rawlins RG & London WT The hemogram, serum biochemistry, and electrolyte profile of aged rhesus monkeys (Macaca mulatta). J. Med. Primatol 12, 184–191 (1983). [PubMed] [Google Scholar]
- 72.Hoffman CL, Higham JP, Mas-Rivera A, Ayala JE & Maestripieri D Terminal investment and senescence in rhesus macaques (Macaca mulatta) on Cayo Santiago. Behav. Ecol 21, 972–978 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kessler MJ, Turnquist JE, Pritzker KP & London WT Reduction of passive extension and radiographic evidence of degenerative knee joint diseases in cage-raised and free-ranging aged rhesus monkeys (Macaca mulatta). J. Med. Primatol 15, 1–9 (1986). [PubMed] [Google Scholar]
- 74.Nussey DH, Froy H, Lemaitre J-F, Gaillard J-M & Austad SN Senescence in natural populations of animals: widespread evidence and its implications for bio-gerontology. Ageing Res. Rev 12, 214–225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bronikowski AM et al. Aging in the natural world: comparative data reveal similar mortality patterns across primates. Science 331, 1325–1328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013). [Google Scholar]
- 77.Testard C et al. Social connections predict brain structure in a multidimensional free-ranging primate society. Sci. Adv 8, eabl5794 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeCasien AR et al. Evolutionary and biomedical implications of sex differences in the primate brain transcriptome. Preprint at bioRxiv 10.1101/2022.10.03.510711 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wong K-S & Pang H-M Simplifying HT RNA quality & quantity analysis. Genet. Eng. Biotechnol. News 33, 17 (2013). [Google Scholar]
- 80.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Warren WC et al. Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science 370, eabc6617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soneson C, Love MI & Robinson MD Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McInnes L & Healy J UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at 10.48550/arXiv.1802.03426 (2018). [DOI] [Google Scholar]
- 85.Schliep KP phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Danecek P et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lipatov M, Sanjeev K, Patro R & Veeramah K Maximum likelihood estimation of biological relatedness from low coverage sequencing data. Preprint at bioRxiv 10.1101/023374 (2015). [DOI] [Google Scholar]
- 92.Alexa A, Rahnenführer J & Lengauer T Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- 93.Alexa A & Rahnenführer J topGO: enrichment analysis for Gene Ontology. Bioconductor 10.18129/B9.bioc.topGO (2019). [DOI] [Google Scholar]
- 94.Durinck S, Spellman PT, Birney E & Huber W Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol 57, 289–300 (1995). [Google Scholar]
- 96.Kang HM et al. Efficient control of population structure in model organism association mapping. Genetics 178, 1709–1723 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Naqvi S et al. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 365, eaaw7317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pletscher-Frankild S, Pallejà A, Tsafou K, Binder JX & Jensen LJ DISEASES: text mining and data integration of disease-gene associations. Methods 74, 83–89 (2015). [DOI] [PubMed] [Google Scholar]
- 99.Wang M et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci. Data 5, 180185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.De Jager PL et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data 5, 180142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Allen M et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci. Data 3, 160089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dunn PK & Smyth GK dglm: double generalized linear models. R package version 1.8.4 https://CRAN.R-project.org/package=dglm (2020). [Google Scholar]
- 104.Herrero J et al. Ensembl comparative genomics resources. Database 2016, bav096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cao J et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cao J et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hennig BP et al. Large-scale low-cost NGS library preparation using a robust Tn5 purification and tagmentation protocol. G3 8, 79–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Korneliussen TS & Moltke I NgsRelate: a software tool for estimating pairwise relatedness from next-generation sequencing data. Bioinformatics 31, 4009–4011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanghøj K, Moltke I, Andersen PA, Manica A & Korneliussen TS Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. Gigascience 8, giz034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Korneliussen TS, Albrechtsen A & Nielsen R ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hedrick PW & Lacy RC Measuring relatedness between inbred individuals. J. Hered 106, 20–25 (2015). [DOI] [PubMed] [Google Scholar]
- 113.Wolock SL, Lopez R & Klein AM Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Darmanis S et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA 112, 7285–7290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu Y et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stuart T et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tasic B et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miller JA et al. Common cell type nomenclature for the mammalian brain. eLife 9, e59928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yao Z et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aran D et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Packer JS et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science 365, eaax1971 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McKenzie AT et al. Brain cell type specific gene expression and co-expression network architectures. Sci. Rep 8, 8868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen L et al. GMPR: a robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ 6, e4600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peters MJ et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun 6, 8570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Friedman J, Hastie T & Tibshirani R Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 126.Anderson JA et al. High social status males experience accelerated epigenetic aging in wild baboons. eLife 10, e66128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study can be downloaded in raw and processed forms from the NCBI Gene Expression Omnibus under series accession number GSE179330.
Datasets/databases used in this study are publicly available and include the rhesus macaque Mmul_10 reference assembly (NCBI RefSeq accession GCF_003339765.1), Ensembl release 101 (http://aug2020.archive.ensembl.org.), KEGG release 100.0 (https://www.genome.jp/kegg), GTEx analysis V8 (dbGaP accession phs000424.v8.p2) and the Alzheimer’s Knowledge Portal AMP-AD meta-analysis (https://doi.org/10.7303/syn11914606).
All requests for biological material should be directed to the CBRU (cbru@pennmedicine.upenn.edu). Requests will be reviewed by the Scientific Review Committee and granted depending on availability, proposed use, investigator funding and other considerations.
All code for this study is accessible through the following GitHub repositories: https://github.com/CayoBiobankResearchUnit/brain_transcriptome_aging_bulk (bulk RNA-seq analysis), https://github.com/CayoBiobankResearchUnit/brain_transcriptome_aging_sc (single-nucleus RNA-seq analysis), https://github.com/bbi-lab/bbi-dmux (sci-RNA-seq3 data demultiplexing) and https://github.com/bbi-lab/bbi-sci. (sci-RNA-seq3 data preprocessing).