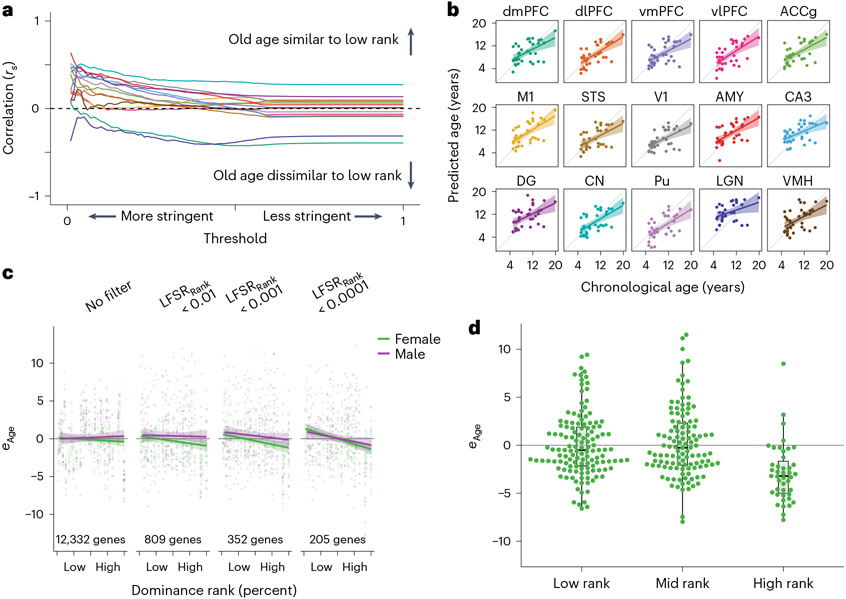
Fig. 6 ∣. Social status signatures of the aging transcriptome.
a, Correlations between age and dominance rank effects robustly increase at more stringent statistical thresholds, indicating similarities between older ages and lower dominance rank. For each threshold, effect estimates were retained when LFSRAge and LFSRRank both passed the threshold on the x axis. Lines correspond with brain regions and follow colors in b. b, An age prediction model robustly predicts chronological age across brain tissue types. All predictions were run using wbaDEG coefficients. Error bands represent the 95% confidence interval of linear model predictions. Prediction results using elastic net (‘glmnet’) models showed similar results (Supplementary Fig. 14). c, Age prediction models filtered to genes with increasingly well-supported dominance rank effects on gene expression reveal stronger support for dominance rank effects on relative transcriptomic age (eAge). Samples from all individuals and brain regions are plotted together. Error bands represent the 95% confidence interval of linear model predictions. d, Comparison of categorical dominance ranks reveals that dominance rank effects on female aging are driven primarily by younger relative ages in high-ranking females. Samples from all individuals and brain regions are plotted together (N = 291 biologically independent samples). Results are shown for predictions filtered first to genes with LFSRRank < 0.001 but were similar across multiple thresholds (Supplementary Fig. 15 and Supplementary Table 37). Box plots depict the median (center) and IQR (bounds of box), with whiskers extending to either the maxima/minima or to the median ± 1.5× IQR, whichever is nearest.