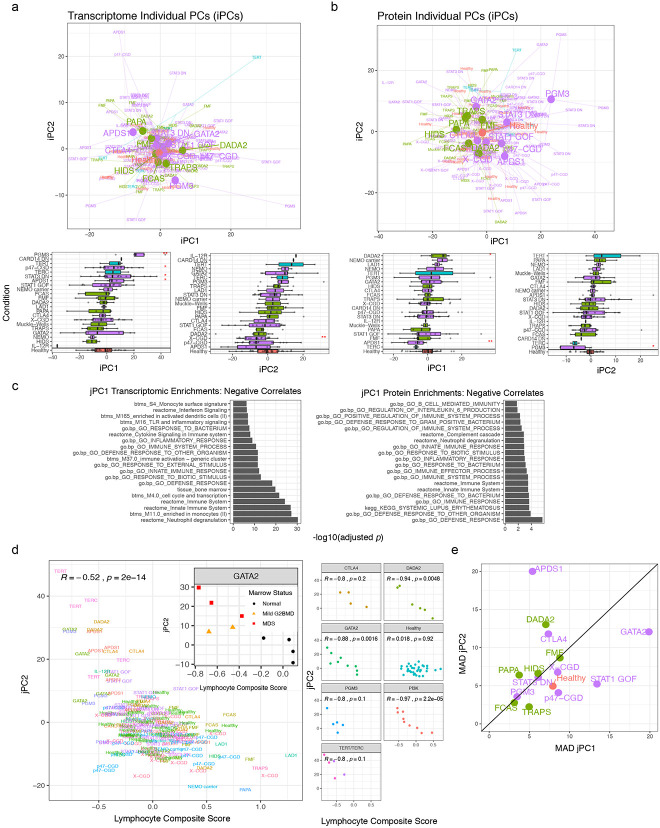
Extended Data Figure 4. Characteristics of the individual and joint PCs from the JIVE analysis.
a, Top panel: patients and healthy subjects shown in transcriptomic individual PC (iPC) 1 vs. iPC2 space. Large dots and text denote the centroid of that disease group. Only conditions with greater than three subjects have a centroid shown. Bottom panels: boxplots of individual transcriptomic iPC1 and iPC2. The rows correspond to the conditions and the color denotes larger condition groups. Box plot center lines correspond to the median value; lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles), and lower and upper whiskers extend from the box to the smallest or largest value correspondingly, but no further than 1.5X inter-quantile range. AI = autoinflammatory diseases. Telo = telomere disorders. PID = primary immunodeficiencies.
b, Similar to (a) but showing the serum protein iPCs.
c, Gene set enrichment of transcriptomic (left) and serum protein (right) features negatively correlated with jPC1 (enrichment calculated using CameraPR; genes/proteins ranked by the Spearman correlation with the JIVE PCs). Gene sets from KEGG pathways, GO biological process gene sets, Reactome pathways, and the blood transcriptomic modules and Human Protein Atlas tissue gene sets.
d, Scatterplot of a hematopoietic composite score (see Methods) vs. jPC2. Left panel displays the trend across all patients including healthy subjects and the right set of panels focus on individual disease groups whose clinical presentation may include marrow failure or lymphopenia. Inset focuses on GATA2 patients, highlighting those with abnormal bone marrow biopsies. Spearman correlation and associated p values are shown. G2BMD = GATA2 deficiency-associated bone marrow disorder. MDS = myelodysplastic syndrome.
e, Scatterplot of Median Absolute Deviation (MAD) of jPC1 and jPC2 scores for each condition in the study. A higher MAD corresponds to greater variation within a disease for that jPC.