Figure 3. lncRNA SLAMR and kinesin KIF5C reciprocally regulate each other.
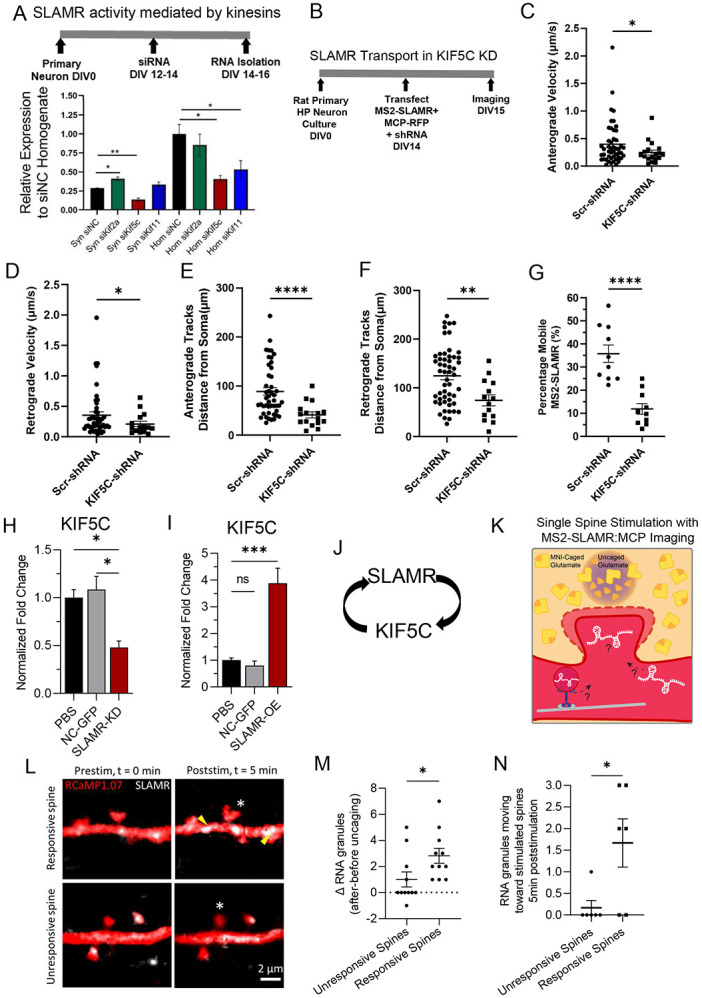
A. Top: Experimental timeline. Bottom: RT-qPCR results show changes in SLAMR RNA levels in total homogenates and synaptic fractions of primary neurons after kinesin silencing. *p<0.05, **p<0.005. One-way ANOVA followed by Dunnett’s test. B. Experimental timeline for KIF5C KD and MS2-SLAMR: MCP-RFP dynamics analysis. C-D. Anterograde and retrograde velocity of MS2-SLAMR in KIF5C knockdown neurons is decreased compared to scrambled control knockdown. Student’s t-test. *p<0.05, Error bars=SEM. N=10 neurons for both conditions. Individual tracks shown. Mean±SEM. E-F. The distance from the soma that anterograde (E) and retrograde tracks (F) of MS2-SLAMR begin their transport is much closer to the soma in KIF5C knockdown neurons compared to scrambled control knockdown. Student’s t-test. **p<0.005, ****p<0.0001. Error bars=SEM. N=10 neurons for both conditions. G. The percent of mobile MS2-SLAMR: MCP-RFP granules is reduced in KIF5C knockdown neurons compared to scrambled control knockdown. Student’s t-test. ****p<0.0001, Error bars=SEM. N=10 neurons for both conditions. H. RT-qPCR results show decreased Kif5c mRNA abundance in neuronal cultures transduced by lentivirus containing shSLAMR compared to NC-GFP lentivirus or treatment with PBS. Student’s t-test *p-value<0.01, **p-value<0.05, ***p-value<0.005. Error bars=SEM. I. RT-qPCR results show increased Kif5c mRNA abundance in neuronal cultures transduced by lentivirus containing OE(over-expressed)-SLAMR compared to NC-GFP or treatment with PBS. Student’s t-test *p-value<0.01, **p-value<0.05, ***p-value<0.005. Error bars=SEM. J. Model of the reciprocal regulation of SLAMR and KIF5C. K. Diagram illustrating single spine stimulation DIV18-21 neurons were transfected with RcAMP1.07 (red) and MS2-SLAMR:MCP (white). 16-22hrs later these neurons were images and individual spines were stimulated with 30 pulses of 2 photon excitation to uncage MNI-Caged glutamate and evaluate transport dynamics of SLAMR to dendritic spines. L. Single frames from spine stimulation experiment prestimulation and 5 minutes after stimulation showing an example of an unresponsive spine which did not increase in volume and a responsive spine which did grow during local glutamate uncaging. Red=RcAMP, White puncta= MS2-SLAMR:MCP granules. White asterisk indicates stimulated spine. M. Results of examining the change in the number of RNA granules (MS2-SLAMR:MCP) before stimulation to 5min after stimulation in a 5μm dendritic region of the stimulated spine. Student’s t-test *p-value<0.05. Error bars=SEM. N. Results of examining the change number of RNA granules (MS2-SLAMR:MCP) moving toward the stimulated spine within 5min after stimulation in a 25μm region of the stimulated spine. Student’s t-test *p-value<0.05. Error bars=SEM.
